Difference between revisions of "Part:BBa K3552015"
(→Characterization) |
|||
| Line 10: | Line 10: | ||
==Characterization== | ==Characterization== | ||
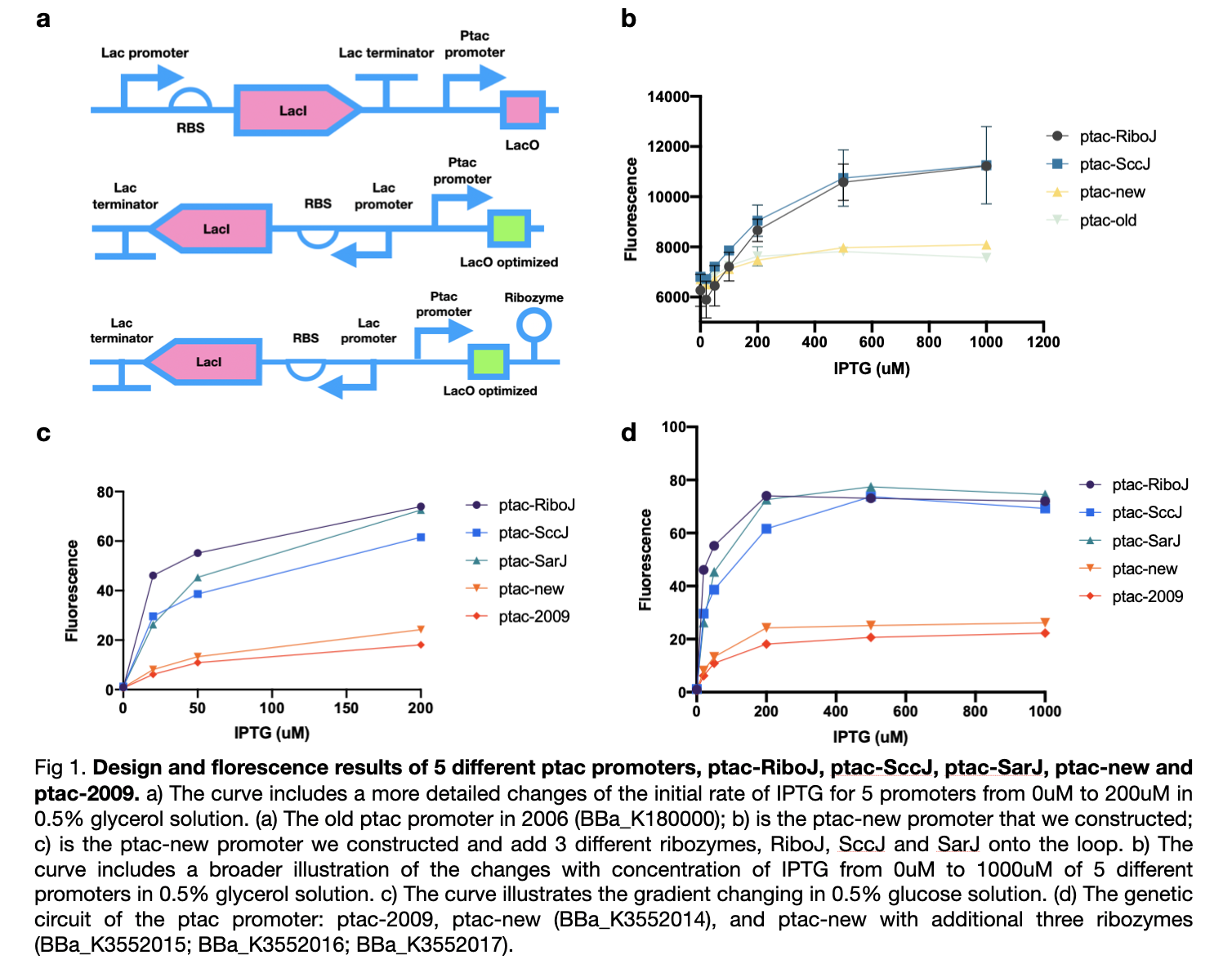
| − | This year we improved the previous ptac promoter, part <partinfo>BBa_K180000</partinfo>, found in 2009 by reversing its lacI promoter, RBS, and terminator which will improve its yield of production. We also added three types of ribozymes, RiboJ, SccJ and SarJ as insulator to prevent the previous effect | + | This year we improved the previous ptac promoter, part <partinfo>BBa_K180000</partinfo>, found in 2009 by reversing its lacI promoter, RBS, and terminator which will improve its yield of production. We also added three types of ribozymes, RiboJ, SccJ, and SarJ as an insulator to prevent the previous effect from the ptac promoter. |
| − | We first carried out an experiment in the culture medium with glucose as the source of carbon for the cultivation of bacteria in M9 medium. In this experiment, we tested four ptac promoter, the old one, the new one, the one with RiboJ and with SccJ. We convinced that all | + | We first carried out an experiment in the culture medium with glucose as the source of carbon for the cultivation of bacteria in the M9 medium. In this experiment, we tested four ptac promoter, the old one, the new one, the one with RiboJ, and with SccJ. We are convinced that all ptac promoter can be activated when induced by IPTG, and as the concentration of IPTG increases, the expression of GFP genes promotes. Furthermore, the addition of two different types of ribozyme shows a prominent improvement of sensitivity towards the concentration of IPTG. At zero concentration of IPTG, all four promoter starts at a similar level, as the concentration of IPTG increases, the gradient of the graph increased dramatically. |
| − | + | Afterward, according to our own medium, which utilizes glycerol as the only source of carbon, we attempted to simulate this situation for all five types of ptac promoter to investigate whether they can fit our experiments or not. Therefore, we substituted the medium to glycerol-M9 medium. The collected results imply that all five promoters can be activated obviously and the same as in glucose, promoters with ribozyme also obtain high sensitivity to the concentration of IPTG and dramatically increased. | |
| + | |||
| + | [[File:T--LINKS_China--Figure 5 different ptac promoter.jpeg|600px]] | ||
These results confirmed the effect of our sensors under various types of carbon sources and maintain nearly no difference in practicability. We modified four new ptac promoter parts which are convinced to have higher sensitivity and adapt better to IPTG than the <partinfo>BBa_K180000</partinfo> discovered in 2009. | These results confirmed the effect of our sensors under various types of carbon sources and maintain nearly no difference in practicability. We modified four new ptac promoter parts which are convinced to have higher sensitivity and adapt better to IPTG than the <partinfo>BBa_K180000</partinfo> discovered in 2009. | ||
| + | |||
| + | ===pili generation=== | ||
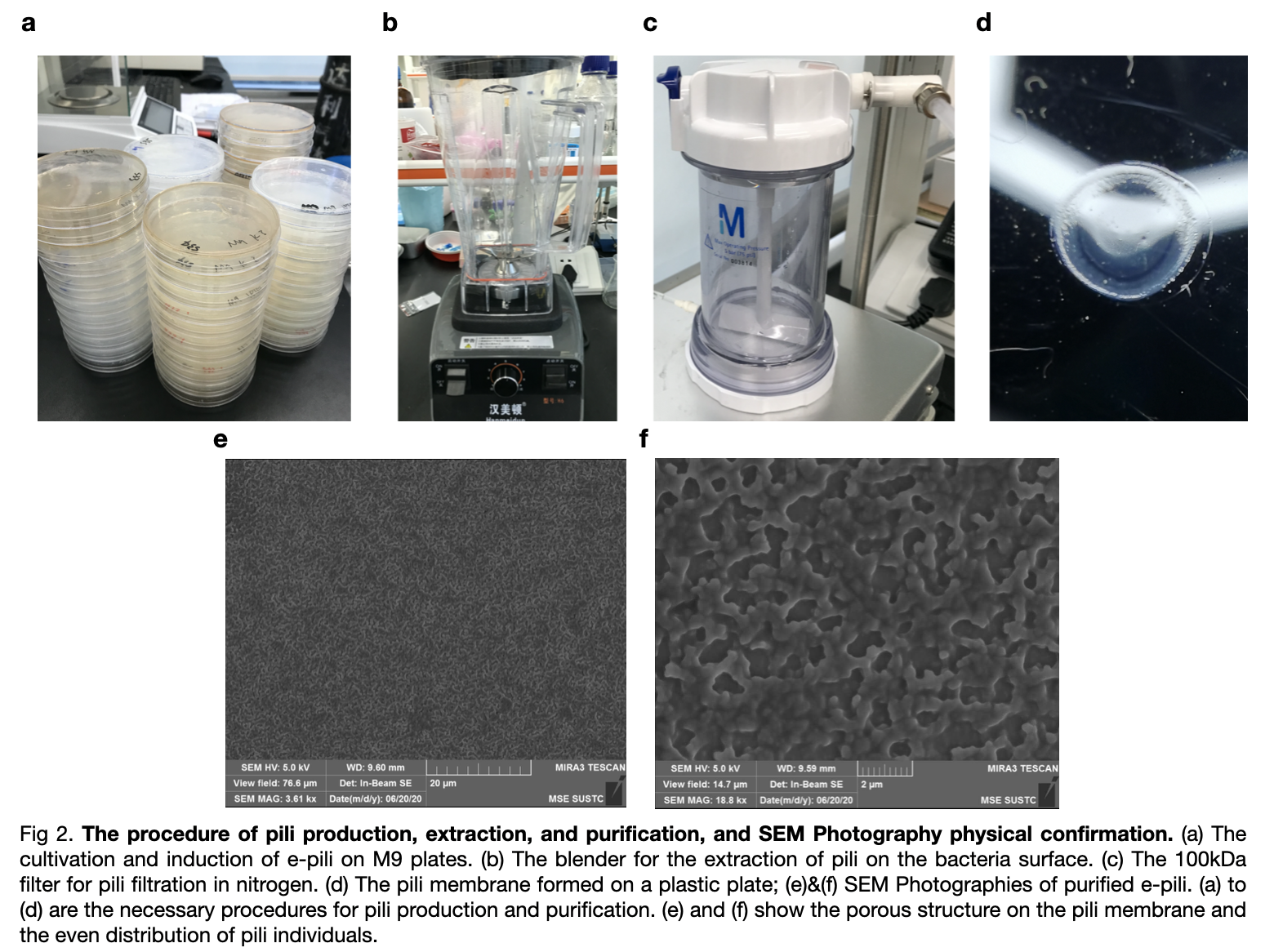
| + | Then we inserted the ptac-RiboJ into our own plasmid instead of the BBa_K180000. The promoter is placed before pilA and we carried out the extraction experiment of pili to verified the function of the ptac-RiboJ promoter. | ||
| + | |||
| + | We transformed the plasmid with ptac-RiboJ promoter into E.coli BL21. Then, cultivated the pili and expressed in vivo. We extracted them finally to 150mM ethanol-amine and filtrate to obtain the required supernatant. We took an SEM photography of the pili which confirm the growth of pilA as there are many hole-like structures in the photography. | ||
| + | |||
| + | [[File:T--LINKS_China--Figure pili extraction.jpeg|600px]] | ||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
Revision as of 14:15, 27 October 2020
Ptac-RiboJ
We improved the part BBa_K180000 to improve its productivity. We reversed the Lac operon parts and changed to a reverse promoter, terminator and RBS. A new ribozyme, RiboJ is added to the sequence. This part is in the part collection where we have 12 genes that code for the base generator of pilA.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Characterization
This year we improved the previous ptac promoter, part BBa_K180000, found in 2009 by reversing its lacI promoter, RBS, and terminator which will improve its yield of production. We also added three types of ribozymes, RiboJ, SccJ, and SarJ as an insulator to prevent the previous effect from the ptac promoter.
We first carried out an experiment in the culture medium with glucose as the source of carbon for the cultivation of bacteria in the M9 medium. In this experiment, we tested four ptac promoter, the old one, the new one, the one with RiboJ, and with SccJ. We are convinced that all ptac promoter can be activated when induced by IPTG, and as the concentration of IPTG increases, the expression of GFP genes promotes. Furthermore, the addition of two different types of ribozyme shows a prominent improvement of sensitivity towards the concentration of IPTG. At zero concentration of IPTG, all four promoter starts at a similar level, as the concentration of IPTG increases, the gradient of the graph increased dramatically.
Afterward, according to our own medium, which utilizes glycerol as the only source of carbon, we attempted to simulate this situation for all five types of ptac promoter to investigate whether they can fit our experiments or not. Therefore, we substituted the medium to glycerol-M9 medium. The collected results imply that all five promoters can be activated obviously and the same as in glucose, promoters with ribozyme also obtain high sensitivity to the concentration of IPTG and dramatically increased.
These results confirmed the effect of our sensors under various types of carbon sources and maintain nearly no difference in practicability. We modified four new ptac promoter parts which are convinced to have higher sensitivity and adapt better to IPTG than the BBa_K180000 discovered in 2009.
pili generation
Then we inserted the ptac-RiboJ into our own plasmid instead of the BBa_K180000. The promoter is placed before pilA and we carried out the extraction experiment of pili to verified the function of the ptac-RiboJ promoter.
We transformed the plasmid with ptac-RiboJ promoter into E.coli BL21. Then, cultivated the pili and expressed in vivo. We extracted them finally to 150mM ethanol-amine and filtrate to obtain the required supernatant. We took an SEM photography of the pili which confirm the growth of pilA as there are many hole-like structures in the photography.