Difference between revisions of "Part:BBa K3629017"
(→Usage and Biology) |
|||
| Line 5: | Line 5: | ||
__TOC__ | __TOC__ | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| + | |||
| + | Fully functional cellulase is composed of: | ||
| + | |||
| + | <ol> | ||
| + | <li>Endoglucanases (EG) which randomly cleave internal beta-bonds of cellulose polymers to make them shorter </li> | ||
| + | <li>Cellobiohydrolases (CBH or exoglucanases) which cleave the shorter polymers to make cellobiose </li> | ||
| + | <ul> | ||
| + | <li>CBHI= Acts on reducing end of sugar molecule </li> | ||
| + | <li>CBHII= Acts on non-reducing end of sugar molecule </li> | ||
| + | </ul> | ||
| + | <li>Beta-glucosidases (BGS) which cleave the cellobiose disaccharide to free glucose units </li> | ||
| + | </ol> | ||
| + | |||
| + | These proteins must be in the correct proportions to each other to efficiently degrade cellulose. | ||
| + | |||
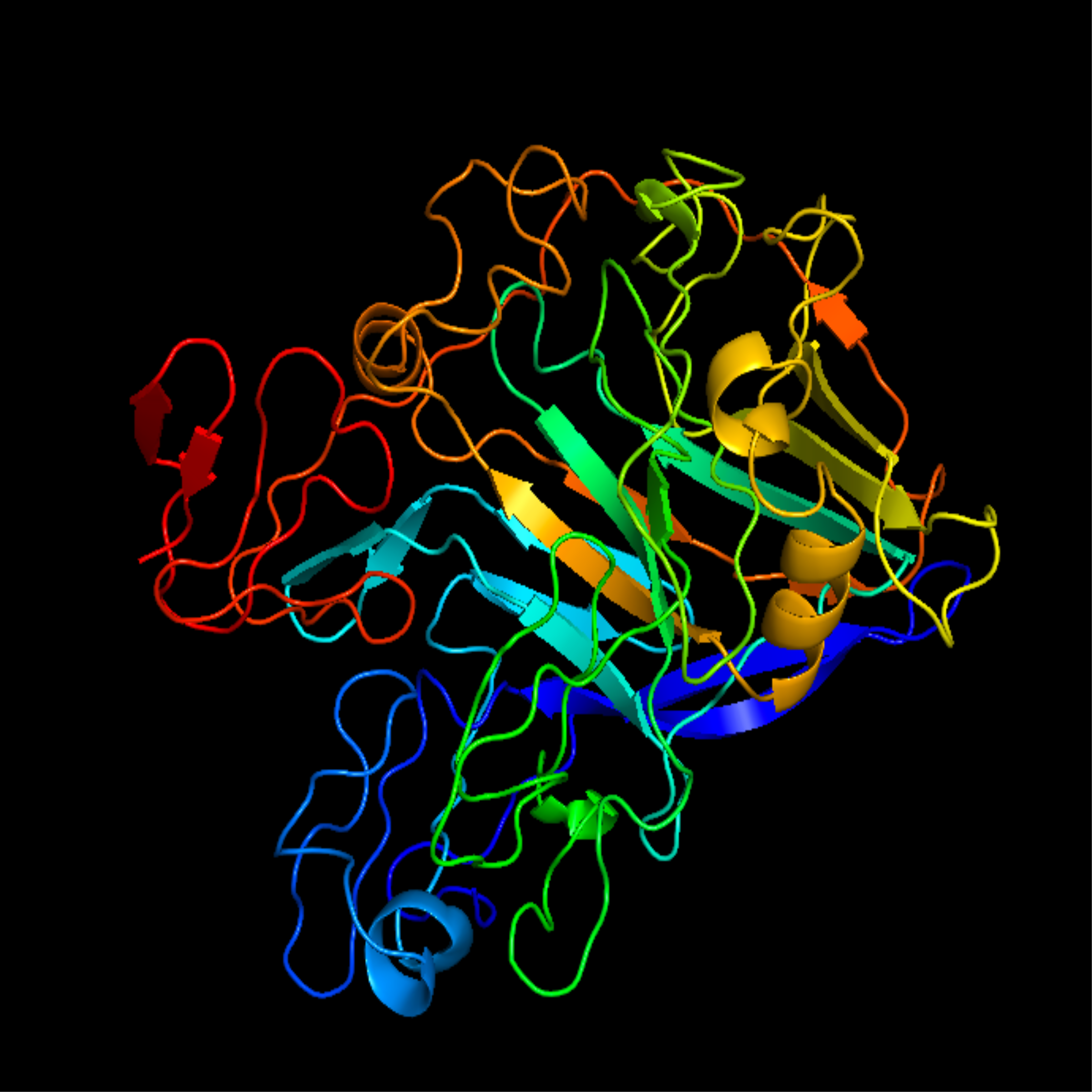
| + | [[Image:T--Calgary--segi.png|250px|thumb|right|Figure 1. Homology model of modified TrEGI]] | ||
| + | |||
| + | This expression construct can be used in the assembly of a EG gene cassette containing Modified TrEGI [https://parts.igem.org/Part:BBa_K3629016 (BBa_K3629016)] and TrEGII [https://parts.igem.org/Part:BBa_K3629017 (BBa_K3629017).] This gene cassette is then intended to be transformed into <i>Y. lipolytica</i> to create a EG- producing strain. This strain should then be co-cultured with two other strains with either a CBH or BGS gene cassette. The three strains together will be able to survive on cellulose media. | ||
===Design=== | ===Design=== | ||
Revision as of 06:36, 27 October 2020
T. reesei EGII expression construct with gibson homology sequences and FLAG tag
Usage and Biology
Fully functional cellulase is composed of:
- Endoglucanases (EG) which randomly cleave internal beta-bonds of cellulose polymers to make them shorter
- Cellobiohydrolases (CBH or exoglucanases) which cleave the shorter polymers to make cellobiose
- CBHI= Acts on reducing end of sugar molecule
- CBHII= Acts on non-reducing end of sugar molecule
- Beta-glucosidases (BGS) which cleave the cellobiose disaccharide to free glucose units
These proteins must be in the correct proportions to each other to efficiently degrade cellulose.
This expression construct can be used in the assembly of a EG gene cassette containing Modified TrEGI (BBa_K3629016) and TrEGII (BBa_K3629017). This gene cassette is then intended to be transformed into Y. lipolytica to create a EG- producing strain. This strain should then be co-cultured with two other strains with either a CBH or BGS gene cassette. The three strains together will be able to survive on cellulose media.
Design
The native signal peptide from T. reesei was removed so it would not interfere with the fused XRP2 secretion tag native to Y. lipolytica.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal prefix found in sequence at 1
Illegal suffix found in sequence at 2835
Illegal EcoRI site found at 2698
Illegal SpeI site found at 749 - 12INCOMPATIBLE WITH RFC[12]Illegal EcoRI site found at 1
Illegal EcoRI site found at 2698
Illegal NheI site found at 68
Illegal NheI site found at 810
Illegal SpeI site found at 749
Illegal SpeI site found at 2836
Illegal PstI site found at 2850
Illegal NotI site found at 7
Illegal NotI site found at 2843 - 21INCOMPATIBLE WITH RFC[21]Illegal EcoRI site found at 1
Illegal EcoRI site found at 2698
Illegal BamHI site found at 2786
Illegal XhoI site found at 125 - 23INCOMPATIBLE WITH RFC[23]Illegal prefix found in sequence at 1
Illegal suffix found in sequence at 2836
Illegal EcoRI site found at 2698
Illegal SpeI site found at 749 - 25INCOMPATIBLE WITH RFC[25]Illegal prefix found in sequence at 1
Illegal EcoRI site found at 2698
Illegal XbaI site found at 16
Illegal SpeI site found at 749
Illegal SpeI site found at 2836
Illegal PstI site found at 2850
Illegal NgoMIV site found at 1496
Illegal NgoMIV site found at 2030
Illegal NgoMIV site found at 2348 - 1000COMPATIBLE WITH RFC[1000]
There is an EcoRI site within the XRP2 terminator, and an SpeI site within the EXP promoter making this part RFC10 incompatible. However, we added the BioBrick prefix and suffix so that the other enzymes (NotI, XbaI, and PstI) could be used to clone this part into an iGEM plasmid or another plasmid. This part can also be cloned through RFC1000 assembly.