Difference between revisions of "Part:BBa K3457048"
| Line 11: | Line 11: | ||
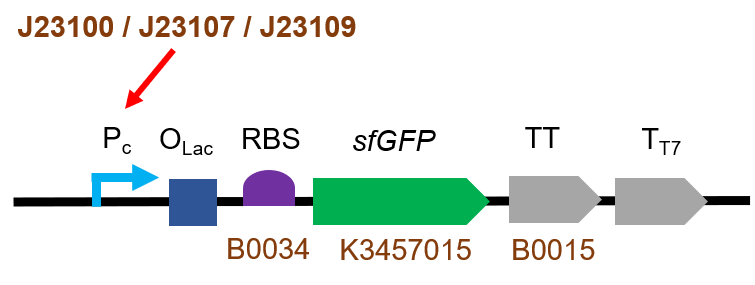
[[File:T--QHFZ-China--100-sfGFP.png|400px|thumb|left|Figure 1. The Schematic cartoon of the DNA construct of BBa_K3457048.]] | [[File:T--QHFZ-China--100-sfGFP.png|400px|thumb|left|Figure 1. The Schematic cartoon of the DNA construct of BBa_K3457048.]] | ||
| − | <p style="clear:left;"> This part was modified from [https://parts.igem.org/Part:BBa_K3457037 BBa_K3457037] T7-Olac-RBS- | + | <p style="clear:left;"> This part was modified from [https://parts.igem.org/Part:BBa_K3457037 BBa_K3457037] T7-Olac-RBS-sfGFP by Golden Gate technique.</p> |
<h3><b>Documentation:</b></h3> | <h3><b>Documentation:</b></h3> | ||
Revision as of 02:19, 27 October 2020
J23100-Olac-RBS-sfGFP
This biological part is the expression sequence of Super-folder Green Fluorescent Protein, also called sfGFP. It was mainly used as the control group of our project this year. We also use it in the experiment to test J23109, J23107 and J23100 promoters.
Contribution
Group: QHFZ-China iGEM 2020
Author: Yixian Yang
Design
This part was modified from BBa_K3457037 T7-Olac-RBS-sfGFP by Golden Gate technique.
Documentation:
Introduction:
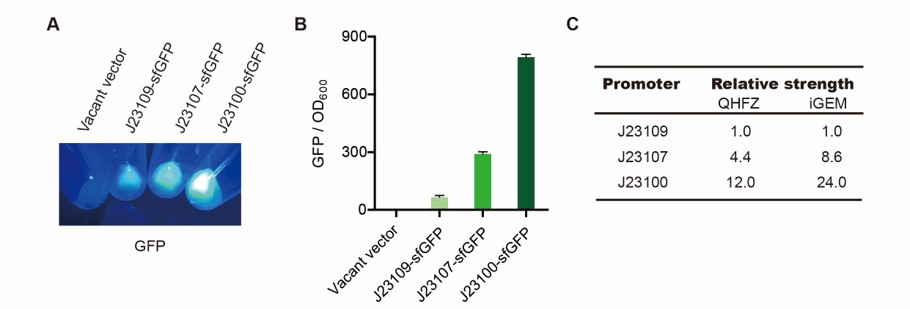
This year, we tried to introduce a new biopreservation method. We used freeze-drying to make the engineered into dry powder. Then the powder can be stored at room temperature for a long time. This method can make the storage of bacteria get rid of ultra-low temperature freezer, so that it will promote the practical application of engineered bacteria out of laboratory. However, the stresses during freeze-drying and subsequent dry storage, including freeze, dry and vacuum, are lethal to bacteria. We use TDPs, to help bacteria survive the situation. In these tests, sfGFP was the control group, as well as a reporter.
Protocol:
We modified a frequently and widely used vector, pet28a+ and put this part into it (Fig. 2). Then we transformed the plasmid into E. coli BL21 strain.
[[File: T--QHFZ-China--J2310-1.png|400px|thumb|left|Figure 2. The Schematic cartoon of the vector.]]
We used the following protocol to detect fluorescence:
(1) Pick clones which are in good condition and put them into 500 μL LB medium containing antibiotics. Shake them to
grow at 37℃ for 5~7 hours until the bacteria solution becomes turbid.
(2) Add 2mM iPTG into 3 mL LB medium containing antibiotics. Add 3 μL of the bacteria solution mentioned in step 1
to dilute the bacteria by the ratio of 1:1000. Shake the solution to grow the bacteria at 37℃ overnight.
(3) The bacteria solution was centrifuged and the LB medium was removed. Then the bacteria was resuspended by PBS.
100 μL such solution was put into a well of a 96-well palte. The GFP fluorescence and OD600 were detected
by a microplate readers (Bio-Teck). The parameters are: exciting light: 488 nm, light reception: 520 nm, gain: 50.
(4) The value of PBS was deducted from the result above. GFP / OD600 was calculated.
Results:
sfGFP was normally expressed. J23100 is a strong promoter.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 11
Illegal NheI site found at 34
Illegal NheI site found at 978 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]