Difference between revisions of "Part:BBa K3338011"
Jonas Scholz (Talk | contribs) (→Cloning) |
Jonas Scholz (Talk | contribs) (→Cloning) |
||
| Line 70: | Line 70: | ||
| − | These primers exhibit 5’ overhangs from approximately 20 bp length, that are designed using SnapGene allowing the assembly via the NEBuilder® HiFi DNA Assembly Cloning Kit. The sequence of the part was verified by sanger sequencing. | + | These primers exhibit 5’ overhangs from approximately 20 bp length, that are designed using SnapGene allowing the assembly via the NEBuilder® HiFi DNA Assembly Cloning Kit. The sequence of the part was verified by sanger sequencing. The vector map of the final construct is shown in figure 1. |
| + | |||
| + | |||
| + | |||
| + | <html> | ||
| + | <center> | ||
| + | <img src="https://2020.igem.org/wiki/images/f/f2/T--Hannover--parts_constD.png" style="width: 50%; height: 50%"> | ||
| + | </p> | ||
| + | Figure 1: Vector map of the CMV-EGFP-MagA-P2A-hGLuc construct in pEGFP-C2. | ||
| + | </center> | ||
| + | </html> | ||
Latest revision as of 00:21, 27 October 2020
CMV-EGFP-MagA-P2A-hGLuc
Usage and Biology
The composite part described here exhibits an EGFP-MagA-P2A-hGLuc-cassette under the control of a CMV-enhancer/promoter for expression in mammalian cells. It was used to determine the expression of MagA and hGLuc at the same time from one promoter.
Cloning
Theoretical Part Design
In order to test whether HeLa cells are able to simultaneously express MagA and hGLuc using a P2A-peptide, MagA and hGLuc were cloned into the pEGFP-C2 vector interspaced with P2A. In the resulting plasmid MagA is encoded with a N-terminal EGFP-tag making MagA detection very easy. The CMV promoter ensures constitutive and high expression rates mimicking the fully activated sensor.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 2553
Illegal BsaI.rc site found at 1885
Illegal BsaI.rc site found at 2434
Illegal SapI site found at 1575
Cloning
The plasmid pEGFP-C2 (BBa_K3338020) was linearized with EcoRI and SalI. The MagA, P2A und hGLuc fragments were PCR amplified using the primers shown in table 1.
| Primer name | Sequence |
|---|---|
| MagA_fw | TACAAGTCCGGCCGGACTCAGATCTCGAGCTCAAGCTTCGccatggacctgcatcatcc |
| MagA_rv | agcaggctgaagttagtagcgattccagtgccaggtccg |
| P2A_fw | ccggacctggcactggaatcgctactaacttcagcctgctgaag |
| P2A_rv | aacagaactttgactcccataggtccagggttctcctcc |
| hGLuc_fw | tggaggagaaccctggacctatgggagtcaaagttctgtttgcc |
| hGLuc_rv | CAGTTATCTAGATCCGGTGGATCCCGGGCCCGCGGTACCGttagtcaccaccggccccct |
These primers exhibit 5’ overhangs from approximately 20 bp length, that are designed using SnapGene allowing the assembly via the NEBuilder® HiFi DNA Assembly Cloning Kit. The sequence of the part was verified by sanger sequencing. The vector map of the final construct is shown in figure 1.
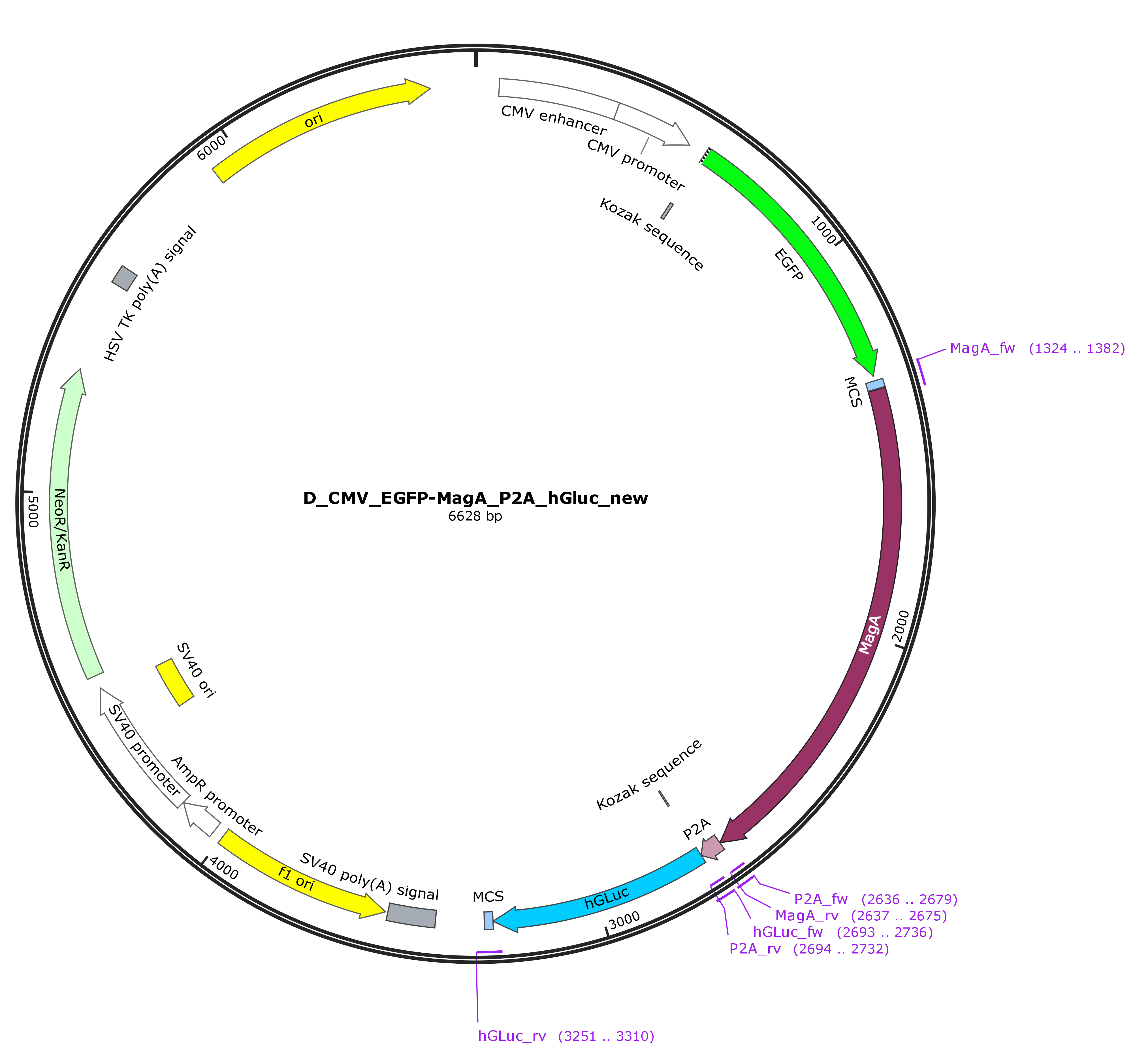 Figure 1: Vector map of the CMV-EGFP-MagA-P2A-hGLuc construct in pEGFP-C2.
Figure 1: Vector map of the CMV-EGFP-MagA-P2A-hGLuc construct in pEGFP-C2.
