Difference between revisions of "Part:BBa K3629003"
(→References) |
(→Design) |
||
| Line 13: | Line 13: | ||
BBa_K3629003 is a unique part in [https://2020.igem.org/Team:Calgary/Part_Collection our collection] and was used in our NpBGS expression construct [https://parts.igem.org/Part:BBa_K3629018 [https://parts.igem.org/Part:BBa_K3629018 (BBa_K3629018)]. We decided to use the TEF1 promoter instead of the TEFin promoter for the BGS part as <i>Y. lipolytica </i> is already thought to express a few endogenous BGSs (1), therefore such high levels of expression are not as required as the other expression constructs in our collection where the TEFin promoter was used in (BBa_K3629012-14, BBa_K36290016, and BBa_K3629023-26). | BBa_K3629003 is a unique part in [https://2020.igem.org/Team:Calgary/Part_Collection our collection] and was used in our NpBGS expression construct [https://parts.igem.org/Part:BBa_K3629018 [https://parts.igem.org/Part:BBa_K3629018 (BBa_K3629018)]. We decided to use the TEF1 promoter instead of the TEFin promoter for the BGS part as <i>Y. lipolytica </i> is already thought to express a few endogenous BGSs (1), therefore such high levels of expression are not as required as the other expression constructs in our collection where the TEFin promoter was used in (BBa_K3629012-14, BBa_K36290016, and BBa_K3629023-26). | ||
| − | |||
This promoter is composed of the wild-type TEF1 promoter (BBa_K2117000) and the first part of the Lip2 signal peptide [https://parts.igem.org/Part:BBa_K1592000 (BBa_K1592000)]. It can be exchanged with the TEFin promoter [https://parts.igem.org/Part:BBa_K3629001 (BBa_K3629001)] placed in our other expression constructs with a Lip2 signal peptide in case of the TEFin promoter not working properly. | This promoter is composed of the wild-type TEF1 promoter (BBa_K2117000) and the first part of the Lip2 signal peptide [https://parts.igem.org/Part:BBa_K1592000 (BBa_K1592000)]. It can be exchanged with the TEFin promoter [https://parts.igem.org/Part:BBa_K3629001 (BBa_K3629001)] placed in our other expression constructs with a Lip2 signal peptide in case of the TEFin promoter not working properly. | ||
| − | |||
[https://parts.igem.org/Part:BBa_K3629012 BBa_K3529012]= <i>T. reesei</i> CBHII expression construct<br> | [https://parts.igem.org/Part:BBa_K3629012 BBa_K3529012]= <i>T. reesei</i> CBHII expression construct<br> | ||
| Line 22: | Line 20: | ||
[https://parts.igem.org/Part:BBa_K3629014 BBa_K3629014]= <i>N. crassa</i> CBHI expression construct <br> | [https://parts.igem.org/Part:BBa_K3629014 BBa_K3629014]= <i>N. crassa</i> CBHI expression construct <br> | ||
[https://parts.igem.org/Part:BBa_K3629016 BBa_K3629016]= Modified <i>T. reesei</i> EGI expression construct <br> | [https://parts.igem.org/Part:BBa_K3629016 BBa_K3629016]= Modified <i>T. reesei</i> EGI expression construct <br> | ||
| − | |||
To do this, the above expression constructs must be digested with BmtI and HindIII to expose the Gibson Y and Gibson | To do this, the above expression constructs must be digested with BmtI and HindIII to expose the Gibson Y and Gibson | ||
| Line 28: | Line 25: | ||
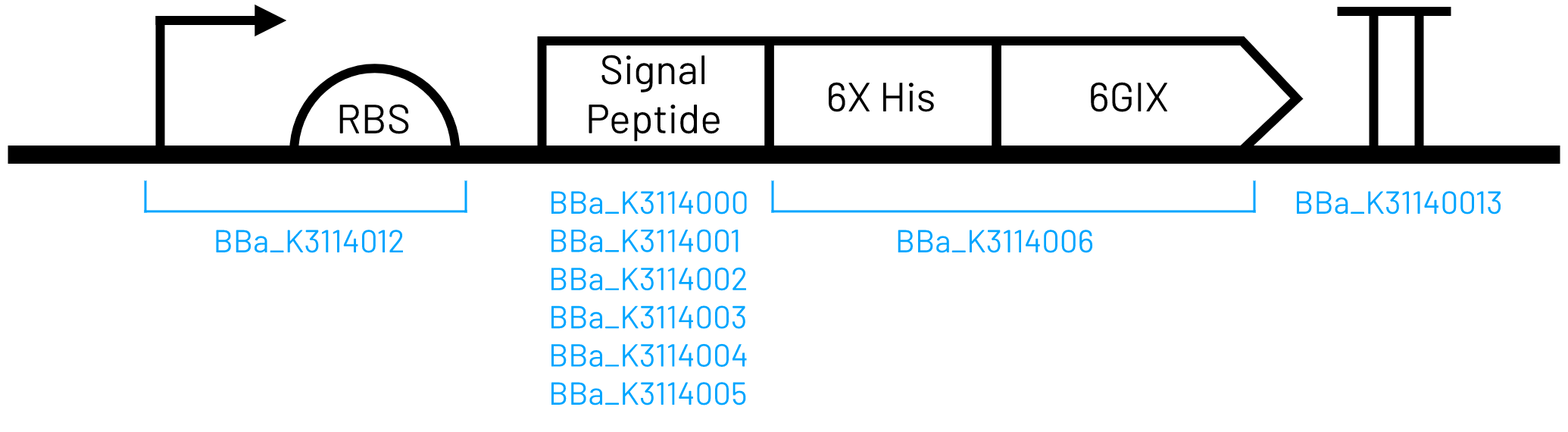
Gibson 1 is used to assemble this promoter with a coding sequence (with the remain 57nts of the Lip2 signal peptide at the N-terminus)into the nourseothricin destination vector [https://parts.igem.org/Part:BBa_K3629015 (BBa_K3629015)]. This coding sequence must have Gibson 2 at the end to ligate into the destination vector successfully (figure 1). | Gibson 1 is used to assemble this promoter with a coding sequence (with the remain 57nts of the Lip2 signal peptide at the N-terminus)into the nourseothricin destination vector [https://parts.igem.org/Part:BBa_K3629015 (BBa_K3629015)]. This coding sequence must have Gibson 2 at the end to ligate into the destination vector successfully (figure 1). | ||
| + | |||
| + | [[Image:T--Calgary--6GIXConstructFinal.png|500px|Figure 1. iGEM Calgary's genetic construct scheme. Each construct consists of a T7 inducible promoter, a strong RBS, one of six signal peptides, the water-soluble chlorophyll binding protein 6GIX with a 6X His tag, and a bidirectional terminator.]] | ||
===Sequence and Features=== | ===Sequence and Features=== | ||
Revision as of 22:29, 26 October 2020
Yarrowia lipolytica TEF1 promoter with first half of Lip2 signal peptide
TEF1 promoter from Yarrowia lipolytica and the first 42nts (14 codons) of the Lip2 signal peptide (BBa_K1592000)from Y. lipolytica.
Usage and Biology
The part contains wild-type TEF1 promoter (nucleotides 1227374 to 1226969) from Yarrowia lipolytica W29 chromosome C (GenBank Accession #CP028450.1) matching BBa_K2117000, and the first 42nts (14 codons) of the Lip2 signal peptide from Y. lipolytica (BBa_K1592000)
Design
BBa_K3629003 is a unique part in our collection and was used in our NpBGS expression construct [https://parts.igem.org/Part:BBa_K3629018 (BBa_K3629018). We decided to use the TEF1 promoter instead of the TEFin promoter for the BGS part as Y. lipolytica is already thought to express a few endogenous BGSs (1), therefore such high levels of expression are not as required as the other expression constructs in our collection where the TEFin promoter was used in (BBa_K3629012-14, BBa_K36290016, and BBa_K3629023-26).
This promoter is composed of the wild-type TEF1 promoter (BBa_K2117000) and the first part of the Lip2 signal peptide (BBa_K1592000). It can be exchanged with the TEFin promoter (BBa_K3629001) placed in our other expression constructs with a Lip2 signal peptide in case of the TEFin promoter not working properly.
BBa_K3529012= T. reesei CBHII expression construct
BBa_K3629013= Modified P. funiculosum CBHI expression construct
BBa_K3629014= N. crassa CBHI expression construct
BBa_K3629016= Modified T. reesei EGI expression construct
To do this, the above expression constructs must be digested with BmtI and HindIII to expose the Gibson Y and Gibson X homology sequences respectively. Meanwhile, this part (BBa_K3629003) must be digested with BbsI to expose the same homology sequences. Once the digested products are placed together in the same tube with the Gibson reagents, the TEFin promoter will be swapped for the TEF1 promoter while maintaining the Lip2 signal peptide for secretion.
Gibson 1 is used to assemble this promoter with a coding sequence (with the remain 57nts of the Lip2 signal peptide at the N-terminus)into the nourseothricin destination vector (BBa_K3629015). This coding sequence must have Gibson 2 at the end to ligate into the destination vector successfully (figure 1).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 53
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
References
1.https://biotechnologyforbiofuels.biomedcentral.com/articles/10.1186/s13068-017-0819-8