Difference between revisions of "Part:BBa K3349011"
Kristiturton (Talk | contribs) |
Kristiturton (Talk | contribs) |
||
| Line 6: | Line 6: | ||
A GFP reporter was added onto the C-terminus. As well a C-terminal his tag was added to the GFP for nickel affinity purification. | A GFP reporter was added onto the C-terminus. As well a C-terminal his tag was added to the GFP for nickel affinity purification. | ||
| + | |||
| + | |||
| + | <html> | ||
| + | <img src= "https://2020.igem.org/wiki/images/c/c9/T--Lethbridge_HS--insertdigest2020.png" alt="pnl modelling" style="width:500px;height:700px;"> | ||
| + | </html> | ||
| + | |||
| + | |||
| + | <b>Figure 1: </b> Restriction digest of PelB and PelC DNA from the pUC57 plasmid using EcoRI and PstI enzymes run on a 1% agarose gel. The red boxes show the band corresponding to that of our DNA constructs. | ||
| + | PelC-GFP in lane 4 shows similar results as well as an unexpected band at around 1500bp. While double checking our construct designs we noticed a illegal EcoRI cut site in the coding sequence that was missed during initial designing. | ||
| + | |||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Revision as of 16:51, 24 October 2020
Pectin Lyase PelC-GFP
The PelC Pectin Lyase is originally from Paenabacillus amylolyticus. This particular pectin lyase is more specific to methylated pectin or homogalacturonan substrates rather than polygalacturonic acid [1]. This protein also works with both PelB (BBa_K3349001) and Pnl (BBa_K3349001) in this organism to effectively break down pectin. PelC has an optimal activity at 55°C and a pH of 10. The Km in in terms of 80% methylated substrates is 0.41 +/-0.06mg/ml. Calcium is a cofactor for this protein and is therefore necessary within the buffer conditions [1].
A GFP reporter was added onto the C-terminus. As well a C-terminal his tag was added to the GFP for nickel affinity purification.
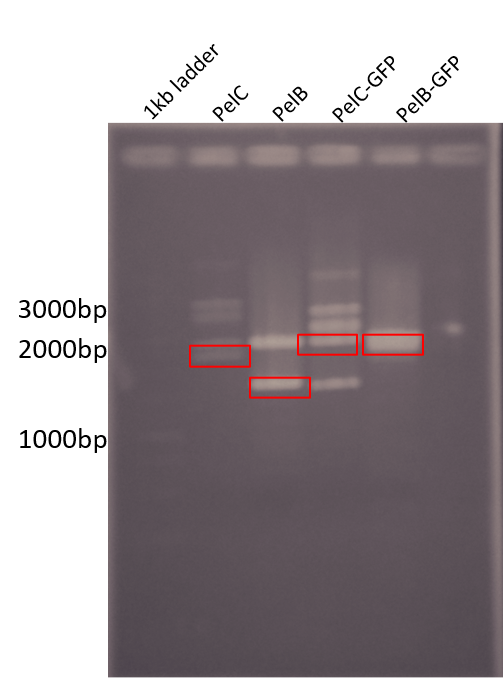
Figure 1: Restriction digest of PelB and PelC DNA from the pUC57 plasmid using EcoRI and PstI enzymes run on a 1% agarose gel. The red boxes show the band corresponding to that of our DNA constructs.
PelC-GFP in lane 4 shows similar results as well as an unexpected band at around 1500bp. While double checking our construct designs we noticed a illegal EcoRI cut site in the coding sequence that was missed during initial designing.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 901
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 287
