Difference between revisions of "Part:BBa K1131000"
(→SZU-China 2020 iGEM TEAM) |
(→SZU-China 2020 iGEM TEAM) |
||
| Line 46: | Line 46: | ||
We used Clustal X to compare the nucleotide and amino acid sequence of FMOfrom the reference with that from BBa_1131000. We found that they have the same amino acid sequence, except for the redundant two amino acids, GS, on the C terminus of bFMO from BBa_1131000. But they have different DNA sequence with 77% identities.<br> | We used Clustal X to compare the nucleotide and amino acid sequence of FMOfrom the reference with that from BBa_1131000. We found that they have the same amino acid sequence, except for the redundant two amino acids, GS, on the C terminus of bFMO from BBa_1131000. But they have different DNA sequence with 77% identities.<br> | ||
<br> | <br> | ||
| − | < | + | <h5>Indigo</h5> |
<br> | <br> | ||
| − | + | The use of indigo can date back to 6,000 years ago. It has been a prevalent dye contributing to the signature tone of blue denim. Indigo have a special way for dying. As indigo is insoluble, it have to be reduced to leuco-indigo for dying. And leuco-indigo can absorbs into fabric while diping and then rapidly oxidized to insoluble indigo after aeration. Thus, indigo can dye cotton without covalent bonds. Indigo as a dye has its own unique properties that’s irreplaceable. While absorbed indigo is robust to detergent for laundering contributing to durability, it is also susceptible to abrasion generating special fraying effect. | |
| − | + | Historically, indigo was extracted form dye-producing plants, such as Indigofera sp.and Polygonum tinctorium. Due to the limited production of indigo extracted from plants, traditional extraction technology was rapidly replaced by the emergent chemical synthethic way in 19th. However, chemical synthetic way of indigo pose a serious threat to the environment, as it need toxic raw materials like aniline derived from petroleum, and hazardous chemical for processing, like reducing agent, formaldehyde, hydrogen cyanide, sodamide, and strong base. Enduring high demand for indigo stimulate the emergence of the much more sustainable way for generating indigo, which appears to be synthesizing heterologous bio-indigo via fermentation. | |
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | 3.2 bFMO Characteristics<br> | ||
| + | 3.2.1 Catalytic Mechanismof FMO<br> | ||
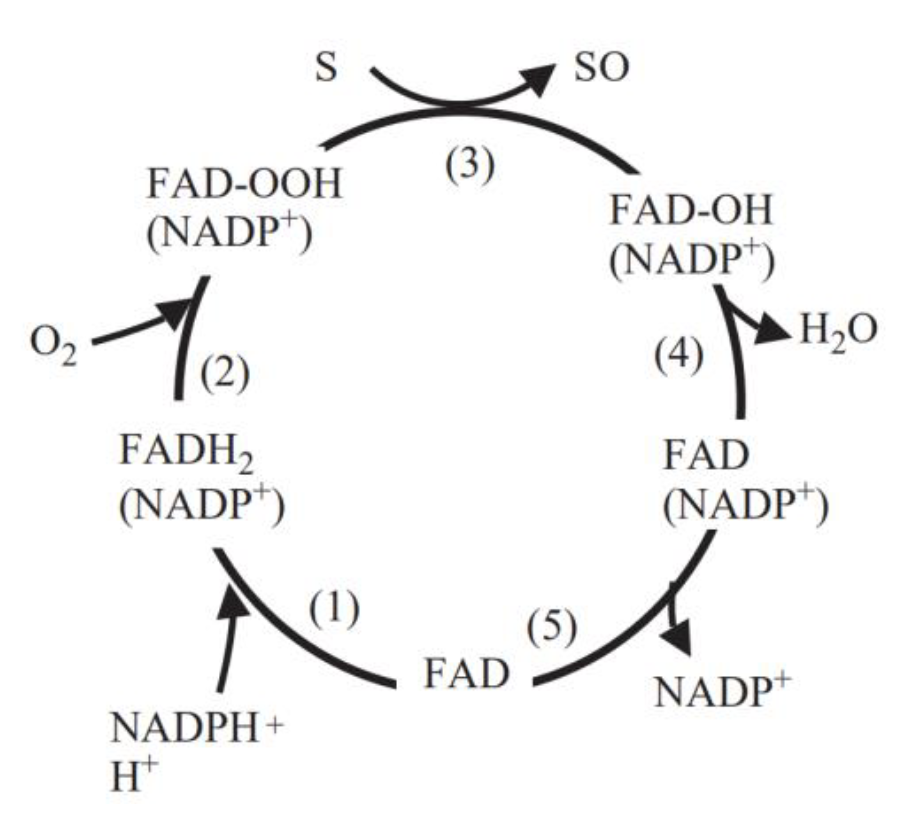
| + | FMO belongs to a class of monooxygenases capable of generating a stable C4a peroxyflavin intermediate. In the first step of the catalytic cycle, FAD undergoes 2-electron reduction by NADPH (Fig. 5). The reduced flavin reacts rapidly with molecular oxygen to form the peroxyflavin and it is in this state, that FMO may exist predominantly in the cell, waiting for a suitable nucleophile with which to react. The nucleophilic attack by the substrate on the FADOOH results in 1 atom of molecular oxygen being transferred to the substrate and 1 atom to form water. The rate-limiting steps in the catalytic cycle are thought to be the breakdown of the FADOH psuedobase or the release of NADP+ . In either case, it is important to note that, unlike the CYP monooxygenase system, substrate binding has no influence on Vmax[2]. | ||
| + | And the most special aspect in this catalytic cycle is that the presence of NADP+ is of critical importance to the stability of the intermediate, and it can keep binding to the enzyme before it finally release at the final step[3]. Binding of NADP can prevent enzyme from being oxidized by NADPH oxidase (Fig. 6). | ||
| + | [[File: Catalytic cycle of flavin-containing monooxygenase.png|center|500px]] | ||
Revision as of 19:37, 18 October 2020
Flavin-containing monooxygenase (FMO); M. aminisulfidivorans
This Flavin-containing monooxygenase (FMO) from M. aminisulfidivorans can be expressed in many strains of E. Coli to produce indigo dye. In the presence of indole and oxygen, FMO can catalyze the addition of a hydroxyl group to indole generating the intermediate indoxyl. Indoxyl then naturally oxidizes to generate indigo which, due to its hydrophobicity, crashes out of solution. The part submitted is the ORF of FMO only.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 1369
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
SZU-China 2020 iGEM TEAM
1.Species Source
In order to better characterize the bacterial flavin-containing monooxygenase (bFMO) documented by BBa_1131000, we found many literature about FMO that are also from M. aminisulfidivorans. The species’current name is Methylophaga aminisulfidivorans MP, and in some early literature it may be named as Methylophaga aminisulfidivorans SK1. Its taxonomy ID in NCBI is 1026882.
2.Experimental Characterization
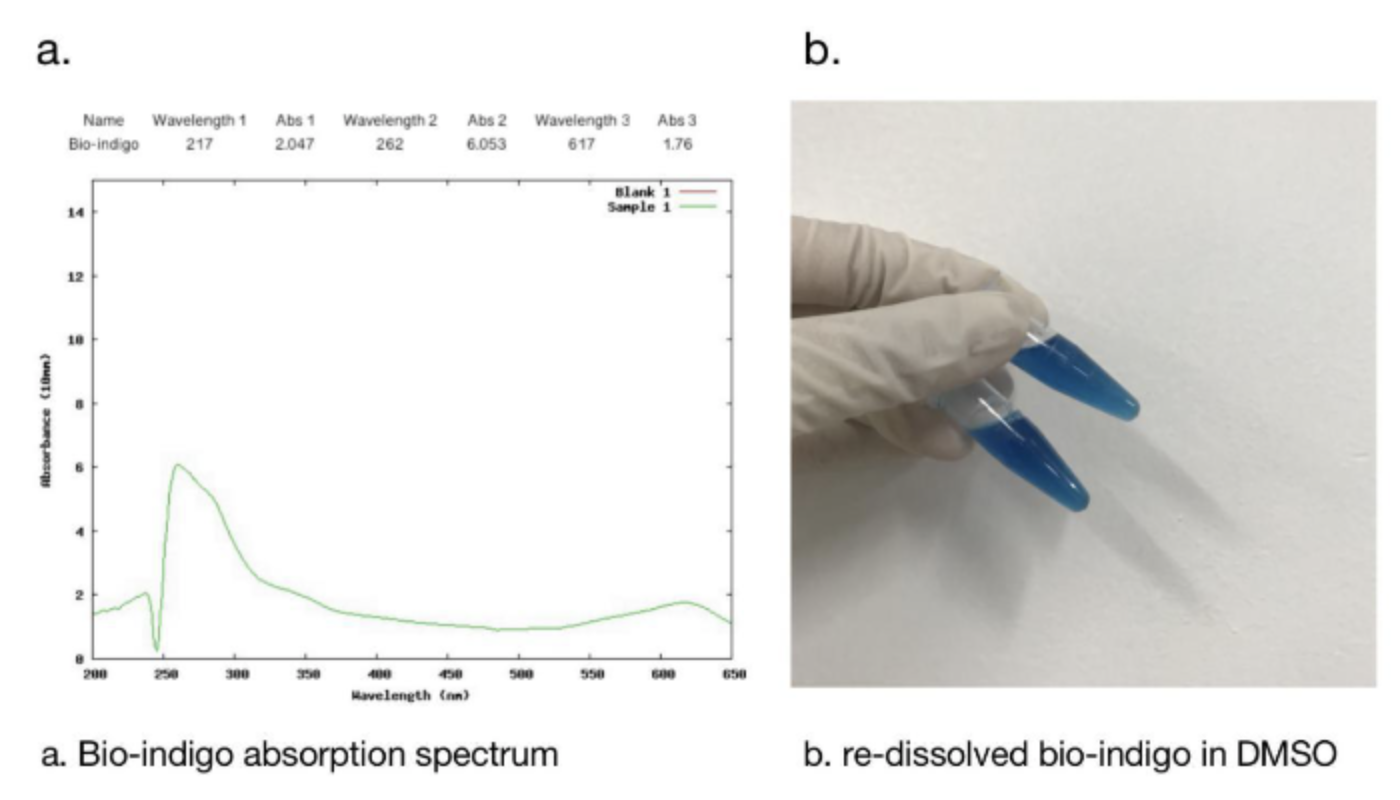
2.1 Bio-indingo Absorption Spectrum
We used DMSO for resolubilization, and bio-indigo had an obvious absorption peak at 617nm, which was the same as that recorded in the literature. And the pigment is clearly blue in daylight.
2.2 Indigo Standard Curve
Prepare indigo standard samples with sample concentrations of 0.8mg/ml, 0.72mg/ml, 0.64mg/ml, 0.48mg/ml, 0.4mg/ml 0.32mg/ml, 0.24mg/ml, 0.16mg/ml, 0.08mg /ml, 0mg/ml, measure the absorbance at 620 nm with an ultraviolet-visible spectrophotometer, and draw a standard curve.
2.3 Determination of Bio-indigo
ProductionThe cells were harvested by centrifugation at 110,000 rpm for 5 mins, after which the supernatant was removed and the precipitation was rinsed by distilled water twice.
Then, the cell pellet was resuspended by dimethyl sulfoxide (DMSO), and tested by ultraviolet-visible spectrophotometer UV-2250 for optical density at 620 nm.
Compare the data obtained in the experiment with the standard curve of the indigo sample to determine the indigo production.
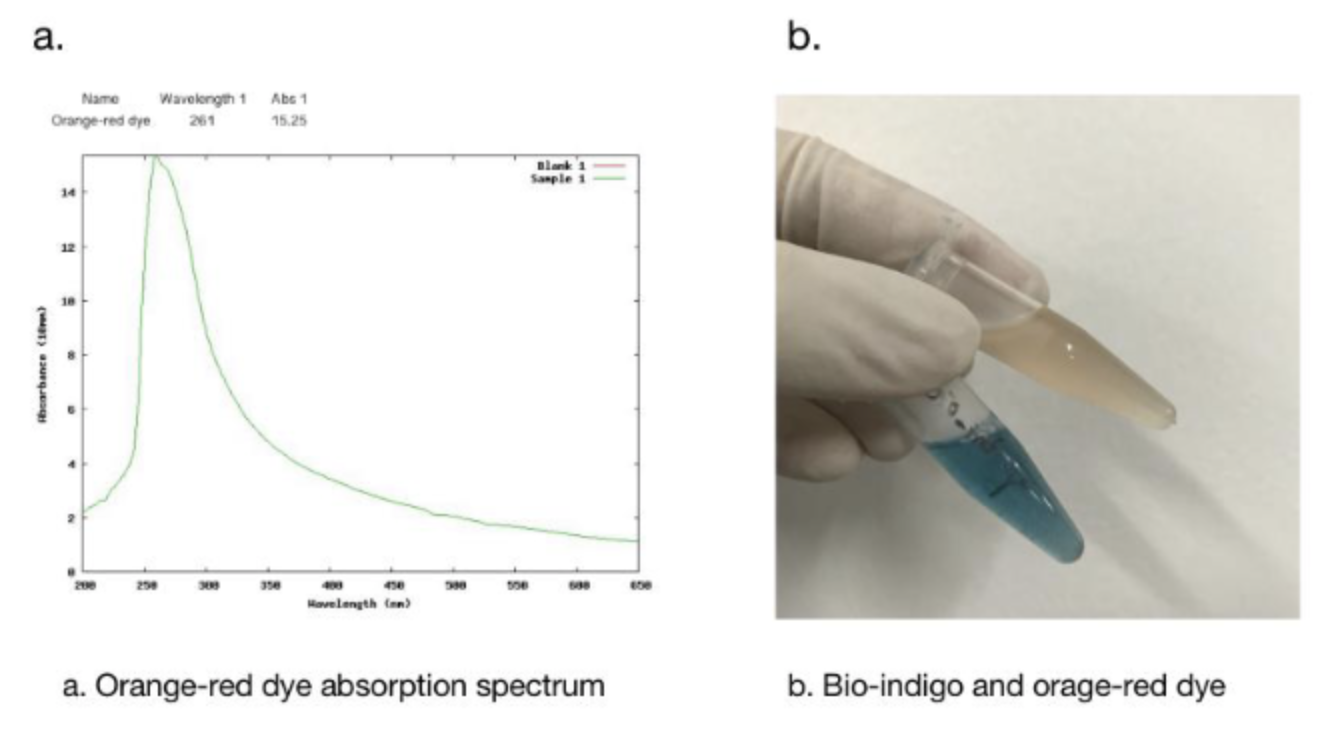
In addition, we got the unexpected orange-red dye in the fermentation product. After we contacted with SHANGHAI_SFLS_SPBS, we found that this phenomenon also happened in their experiments. After consulting the literature, we found that this may be due to the production of isatin, which is obtained by oxidation of indigo. It is preliminarily speculated that this is due to the overexpression of FMO, which caused the indigo to be further oxidized into isatin.
3.Documentary Characterization
3.1 Background
FMO
Flavin-containing monoocygenases (FMOs) are first discovered in rat liver microsomes, and are usually found in eukaryotes. The amount of enzyme present in different tissues varies with species and sex, but the highest concentration is usually found in the liver. FMOs are involved in a wide range of oxidative biological processes, including drug detoxification, xenobiotic metabolism and bio-catalytic synthesis by catalyzation of the oxygenation of many nitrogen-, sulfur-, phosphorous-, selenium-, and other nucleophilic heteroatom.
In the literature in 2003, Choi, H.S.et alcloned the first discovered bacterial FMO from a methylotrophic bacterium they isolated from the seawater at Mokpo, Korea [1]. And they named the new species Methylophaga aminisulfidivorans SK1. It grows on methanol, methylated amines, and dimethylsulfide but not on methane. Fructose and glucose are the only multicarbon compounds tested that can be used as growth substrates.
They isolated and cloned a gene of Methylophaga aminisulfidivorans SK1whose presence in E.coli produced blue dye indigo. The deduced amino acid sequence from the gene showed approximately 30% identities with FMOs of human (FMO1-FMO5). Its biochemical properties such as substrate specificities and absorption spectra were similar to the eukaryotic FMO families. Thus, Hack Sun Choi et al assigned the enzyme to be a bacterial FMO. And the nucleotide sequence of the bacterial fmo gene has been deposited in the GenBank database under Accession No. AF494423.
We used Clustal X to compare the nucleotide and amino acid sequence of FMOfrom the reference with that from BBa_1131000. We found that they have the same amino acid sequence, except for the redundant two amino acids, GS, on the C terminus of bFMO from BBa_1131000. But they have different DNA sequence with 77% identities.
Indigo
The use of indigo can date back to 6,000 years ago. It has been a prevalent dye contributing to the signature tone of blue denim. Indigo have a special way for dying. As indigo is insoluble, it have to be reduced to leuco-indigo for dying. And leuco-indigo can absorbs into fabric while diping and then rapidly oxidized to insoluble indigo after aeration. Thus, indigo can dye cotton without covalent bonds. Indigo as a dye has its own unique properties that’s irreplaceable. While absorbed indigo is robust to detergent for laundering contributing to durability, it is also susceptible to abrasion generating special fraying effect.
Historically, indigo was extracted form dye-producing plants, such as Indigofera sp.and Polygonum tinctorium. Due to the limited production of indigo extracted from plants, traditional extraction technology was rapidly replaced by the emergent chemical synthethic way in 19th. However, chemical synthetic way of indigo pose a serious threat to the environment, as it need toxic raw materials like aniline derived from petroleum, and hazardous chemical for processing, like reducing agent, formaldehyde, hydrogen cyanide, sodamide, and strong base. Enduring high demand for indigo stimulate the emergence of the much more sustainable way for generating indigo, which appears to be synthesizing heterologous bio-indigo via fermentation.
3.2 bFMO Characteristics
3.2.1 Catalytic Mechanismof FMO
FMO belongs to a class of monooxygenases capable of generating a stable C4a peroxyflavin intermediate. In the first step of the catalytic cycle, FAD undergoes 2-electron reduction by NADPH (Fig. 5). The reduced flavin reacts rapidly with molecular oxygen to form the peroxyflavin and it is in this state, that FMO may exist predominantly in the cell, waiting for a suitable nucleophile with which to react. The nucleophilic attack by the substrate on the FADOOH results in 1 atom of molecular oxygen being transferred to the substrate and 1 atom to form water. The rate-limiting steps in the catalytic cycle are thought to be the breakdown of the FADOH psuedobase or the release of NADP+ . In either case, it is important to note that, unlike the CYP monooxygenase system, substrate binding has no influence on Vmax[2].
And the most special aspect in this catalytic cycle is that the presence of NADP+ is of critical importance to the stability of the intermediate, and it can keep binding to the enzyme before it finally release at the final step[3]. Binding of NADP can prevent enzyme from being oxidized by NADPH oxidase (Fig. 6).