Difference between revisions of "Part:BBa K3292004"
| Line 14: | Line 14: | ||
To characterize this improvement we first performed a fluorescence assay in order to observe the production of this protein by induction with IPTG | To characterize this improvement we first performed a fluorescence assay in order to observe the production of this protein by induction with IPTG | ||
at different concentrations. In addition, we performed an SDS-PAGE to observe our expressed protein under the same induction conditions by different concentrations of IPTG. | at different concentrations. In addition, we performed an SDS-PAGE to observe our expressed protein under the same induction conditions by different concentrations of IPTG. | ||
| − | + | ||
<div class = "left"> | <div class = "left"> | ||
Latest revision as of 02:50, 22 October 2019
6His-tagged Super Folding GFP with terminator
This part is an improvement of the BBa_I746916 which allows you to choose which is the better option for your protein either to purify it with an affinity column or just to take it from the medium.
Improvement of BBa_I746916 by iGEM TecMonterrey
The BioBrick BBa_I746916 corresponds to the coding sequence for the sfGFP. According to Zhang et al. (2019), this protein presents an auto-secretory activity, and can also secrete other proteins when linked to them. Based on this, we decided to improve this part by adding a 6x His-tag to facilitate other teams the recovery of proteins of interest, and compare it with other traditional purification methods.
To characterize this improvement we first performed a fluorescence assay in order to observe the production of this protein by induction with IPTG at different concentrations. In addition, we performed an SDS-PAGE to observe our expressed protein under the same induction conditions by different concentrations of IPTG.
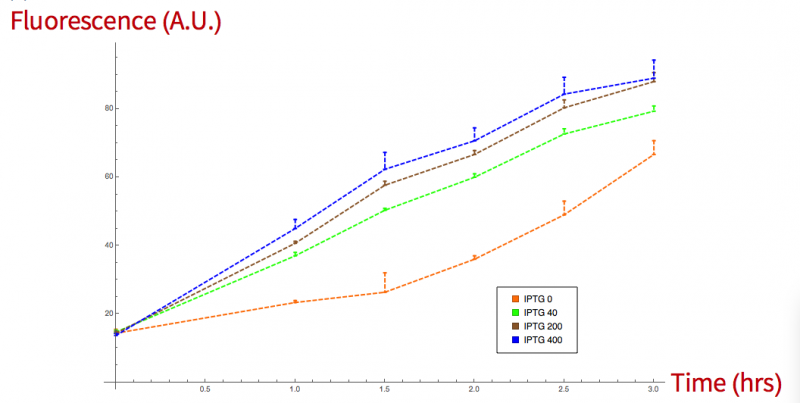
Figure 1 (Fluorescence essay) Fluorescence vs time of sfGFP production induced at 0, 40, 200 and 400μM.

Figure 2 (SDS-Page). SDS-PAGE from left to right the samples are, the lysate of an induced E. Coli producing the sfGFP, 20mM Imidazole fraction of Ni=NTA purification,
sfGFP 150mM Imidazole elution fraction, supernatant from sfGFP producer, negative control (an E. Coli transformed with a Psb1c3 only containing a promoter between
the prefix and suffix), sialidase induced supernatant and non induced supernatant, and an sfGFP non induced. Induction was done at 400μM.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 13
