Difference between revisions of "Part:BBa K3190501"
| Line 33: | Line 33: | ||
<b> Yeast transformation: genome integration </b> | <b> Yeast transformation: genome integration </b> | ||
| − | + | Initially our yeast was transformed with an episomal plasmids containing BAX under a galactose inducible promoter. For our control strain we used an empty vector. Following transformation, each culture were split into two. One half was plated on a plate containing raffinose selection media with 0.0 % galactose, and the other half on raffinose selection media containing 1.0 % galactose. In both strains a second empty vector was introduced, placing both strains under Uracil (U) and Tryptophan (W) selection. | |
| − | + | As can be seen on Figure 2, no colonies were seen on the plate containing galactose, suggesting that the BAX protein was expressed in such a high concentration that all cells died. For our control strain an equal amount of cells were seen both with and without galactose. | |
| + | |||
| + | For more stable expression, we also interated pGAL1-BAX into the chromosome of <i>S. cerevisiae</i>, using U selection. Here, positive transformants were selected for by performing yeast colony PCR (Figure 1). This strain was used for all further experiments. | ||
| + | |||
| + | [[File:ovulaid29.jpg|300px]] | ||
| + | |||
| + | <small> <b> Figure 1: Yeast colony PCR of genomically integrated transformants</b> | The positive colony of yeast is confirmed by the expected band size of around 800 bp.</small> | ||
| + | |||
| + | [[File:PGAL1-BAX A.jpeg|300px]] [[File:PGAL1-BAX B.jpeg|300px]] | ||
| + | |||
| + | <small><b>Figure 2: Transformant plates of dual plasmid transformed <i>S. cerevisiae</i></b> | Both transformed with pGAL1-BAX (URA marker) and an empty vector (TRP marker). A: plate with no galactose. B: plate with 1 % galactose. </small> | ||
| − | |||
| − | |||
<b> Yeast transformation: dual plasmid transformation </b> | <b> Yeast transformation: dual plasmid transformation </b> | ||
Revision as of 22:56, 21 October 2019
BAX: Bcl-2-associated X protein CDS
Coding sequence of the mammalian apoptosis regulator Bax protein. The gene will be integrated into the genome of the fungal chassis Saccharomyces cerevisiae, acting as the key gene in our biosafety device. Overexpression of Bax is lethal in S. cerevisiae, leading to the release of cytochrome c from mitochondria and thus apoptosis. The following results are characterization of a part submitted by iGEM17_NAU-CHINA: BBa_K2365048.
Usage and Biology
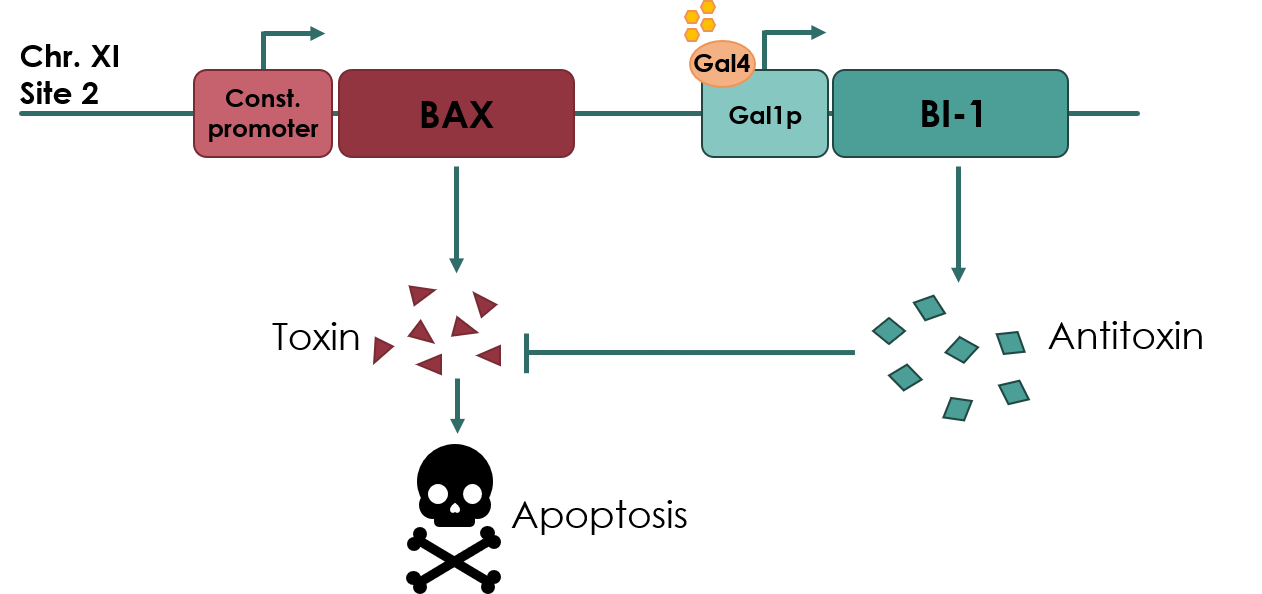
For our biosafety device, the Bax protein will be under the expression of a constitutive promoter, pADH2 (No part name specified with partinfo tag.). An anti-toxin, BAX Inhibitor-1 (BI-1) (BBa_K3190502) will be under the expression of an inducible promoter, pGAL1 (BBa_K3190050). As long as there is galactose present in the media, the anti-toxin will be expressed, inhibiting the apoptotic effects of Bax protein. If the cell escapes the media, the anti-toxin will no longer be expressed, and Bax protein will cause apoptosis of the cell.
Figure legend: Overview of our toxin/anti-toxin biosafety device.
Designing the construct
In order to characterize the Bax protein, we cloned it to inducible promoter pGAL1 (BBa_K3190050), as expression under a constitutive promoter would be expected to cause the cells to die.
Using USER ligation, we assembled the BAX gene with pGAL1 on a plasmid backbone compatible with multiplex integration cassette. The backbone used contains a URA selection marker, and will integrate the construct in the yeast genome at chromosome 11, site 2.
E. coli cloning of pGAL1-BAX
The construct was successfully cloned in E. coli as confirmed by below gel image of colony PCR. We used forward primer for the promoter (pGAL1) and reverse primer for the gene (BAX). We therefore expect a band of 1034 bp.
[INSERT GEL IMAGE!!] Figure Legend: Above gel electrophoresis image shows the positive colony PCR sample. A band was observed around 1000 bp, which correlates well to the expected band size for a construct of 1034 bp.
In order to further characterize the expression of BAX under pGAL1, we also transformed yeast with a dual plasmid system. Using USER ligation, we assembled pGAL1 and BAX on a high copy plasmid backbone containing a URA selection marker. The construct was successfully cloned in E. coli, as depicted by below colony PCR gel image:
[Insert pUUS gel image] Figure legend: Gel image showing the positive E. coli colonies of pGAL1-BAX. The expected band is seen around 1000 bp.
Yeast transformation: genome integration
Initially our yeast was transformed with an episomal plasmids containing BAX under a galactose inducible promoter. For our control strain we used an empty vector. Following transformation, each culture were split into two. One half was plated on a plate containing raffinose selection media with 0.0 % galactose, and the other half on raffinose selection media containing 1.0 % galactose. In both strains a second empty vector was introduced, placing both strains under Uracil (U) and Tryptophan (W) selection.
As can be seen on Figure 2, no colonies were seen on the plate containing galactose, suggesting that the BAX protein was expressed in such a high concentration that all cells died. For our control strain an equal amount of cells were seen both with and without galactose.
For more stable expression, we also interated pGAL1-BAX into the chromosome of S. cerevisiae, using U selection. Here, positive transformants were selected for by performing yeast colony PCR (Figure 1). This strain was used for all further experiments.
Figure 1: Yeast colony PCR of genomically integrated transformants | The positive colony of yeast is confirmed by the expected band size of around 800 bp.
Figure 2: Transformant plates of dual plasmid transformed S. cerevisiae | Both transformed with pGAL1-BAX (URA marker) and an empty vector (TRP marker). A: plate with no galactose. B: plate with 1 % galactose.
Yeast transformation: dual plasmid transformation
We picked the positive E. coli colonies of the pGAL1-BAX cloned in a high copy plasmid, and purified the DNA from these. After confirming the sequence, we transformed the construct into S. cerevisiae. To select for positive transformants, we also transformed an empty vector with a TRP selection marker, and grew the colonies on plates without both URA and TRP. As a control, we spread the cells on plates both with and without galactose. No growth was detected in the galactose containing plate, as seen below:
[Insert plate image of yeast 10] Figure legend: Transformant plates of dual plasmid transformed S. cerevisiae, transformed with pGAL1-BAX and an empty vector. Right: plate with 1 % galactose. Left: plate with no galactose.
Galactose induction assay To compare
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 452
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]