Difference between revisions of "Part:BBa K3113302"
Theresakeil (Talk | contribs) |
Theresakeil (Talk | contribs) |
||
| Line 41: | Line 41: | ||
<html> | <html> | ||
<figure class="figure"> | <figure class="figure"> | ||
| − | <img src="https://2019.igem.org/wiki/images/ | + | <img src="https://2019.igem.org/wiki/images/1/1e/T--Munich--VLP_DLS_HEK_empty_vs_loaded.png" width="400" height="360" class="figure-img img-fluid rounded" alt="Size distribution frequency of VLPs in HEK293T"> |
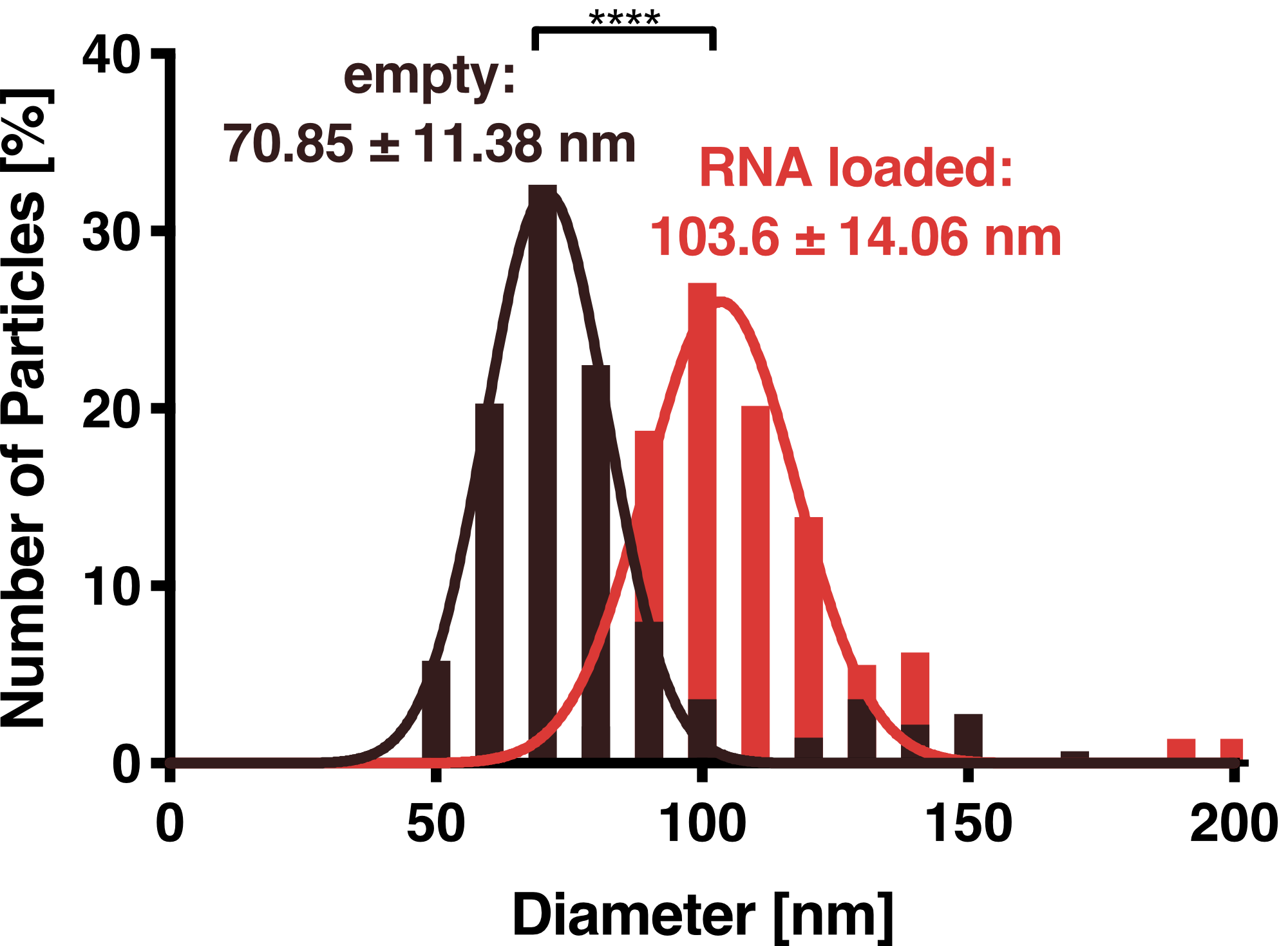
| − | <figcaption class="figure-caption"><b> | + | <figcaption class="figure-caption"><b>Figure 2: Dynamic light scattering (DLS) measurement of VLPs.</b> Purified VLPs either with or without the adapter protein L7Ae were measured. Unloaded and loaded vesicles showed a narrow size distribution with mean diameters of 71 and 104 nm, respectively. |
| − | Figure | + | |
</figcaption> | </figcaption> | ||
</figure> | </figure> | ||
</html> | </html> | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
Revision as of 22:45, 21 October 2019
pCAG_Gag-HiBiT-L7Ae
This plasmid codes for the coat protein of gag fused to a split luciferase fused to an RNA binding protein. This construct forms virus-like particles which can be detected via a split luciferase assay. Through the RNA binding protein, the vesicles can be loaded with RNA.
Usage
pCAG_GAG-HiBit-L7Ae construct is a Composite part that has been designed to achieve VLP formation inside HEK293T/MIN6-K8 cells and enable the transport of chosen RNA information inside vesicles through L7Ae-C/D box interaction (together with construct BBa_K3113301 and BBa_K3113303).
Biology
Gag-p24 is the group specific antigen from the lentivirus HIV. It assembles at the plasma membrane and leads to the budding of vesicles.[1]
“The Nano-Glo® HiBiT Lytic Detection System quantifies cellular protein in minutes with high sensitivity and a broad dynamic range using a simple add-mix-read assay format.”[2] The HiBiT peptide is an 11 amino acid long tag that can be added to the protein of interest. The tagged protein can be measured with the Nano-Glo® HiBiT Detection Reagent which contains the LbBiT. Together the HiBiT and the LgBiT form a functioning luminescent NanoBiT® enzyme.
The archaeal L7Ae and eukaryotic 15.5kD protein homologs are members of the L7Ae/15.5kD protein family that characteristically recognize K-turn motifs found in both archaeal and eukaryotic RNAs. In Archaea, the L7Ae protein uniquely binds the K-loop motif found in box C/D.[3]
The k-turn is a ubiquitous structural motif in RNA forming a very tight kink in the axis of helical RNA that plays an important role in many aspects of RNA function. L7Ae is a member of a superfamily of proteins that bind k-turns in RNA, stabilizing the tightly kinked conformation. They are extremely widespread and are important in the assembly of RNA–protein complexes central to translation, splicing and site-specific RNA modification. [4]
Characterization
Transmission Electron Microscopy (TEM)
To validate that our vesicles are intact and properly shaped, we prepared purified VLPs and exosomes for transmission electron microscopy (TEM). This microscopy technique is based on a high-energy beam of electrons shown through a very thin sample fixed on a grid and allows high-resolution imaging.
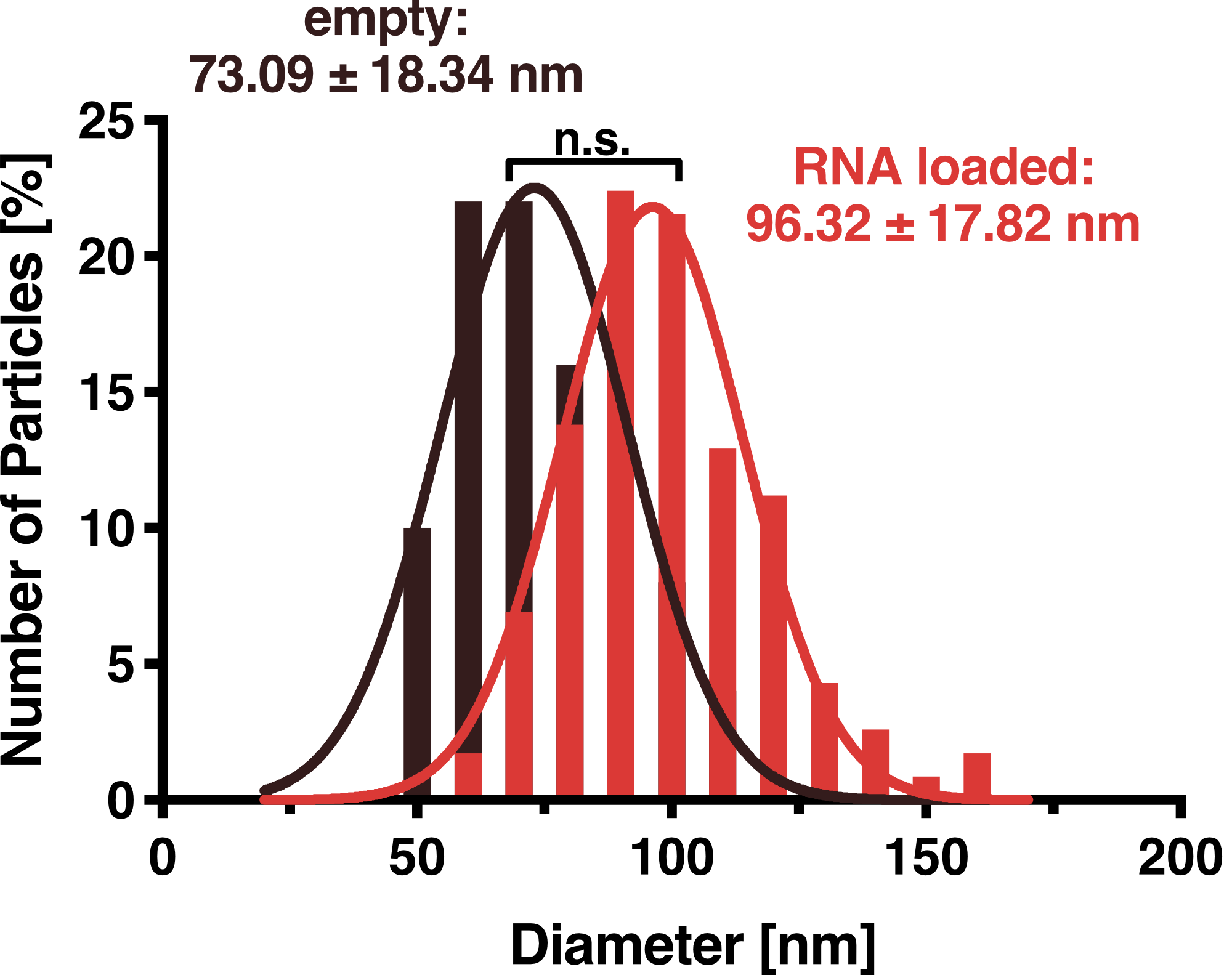
Dynamic Light Scattering (DLS)
To further characterize our vesicles and determine the size distribution and sample homogeneity, we performed Dynamic Light Scattering (DLS) and Transmission Electron Microscopy (TEM) to determine the size and shape of our vesicles.
DLS measurements of VLP particles showed a narrow Gaussian size distribution indicating that the samples are very homogenous. Interestingly, a shift of about 30 nm is seen between cargo-loaded and unloaded VLPs; cargo-loaded particles have a mean diameter of 104 ± 14 nm, whereas unloaded vesicles showed a mean diameter of 71 ± 11 nm.
Split-luciferase bioluminescent assay: The HiBiT Assay
To prove that the BioBrick Part we designed works as expected, we performed a HiBit split-luciferase assay, which shows luminescent signal detected in fully formed VLPs. On the graph below the data shows that engineered Gag protein has been expressed in HEK293T cell line. Further on, based on this data, we have calculated the export to be in approx. 50% for transfection with as well as without adapter construct. With this, we report that Gag has successfully assembled into VLPs.

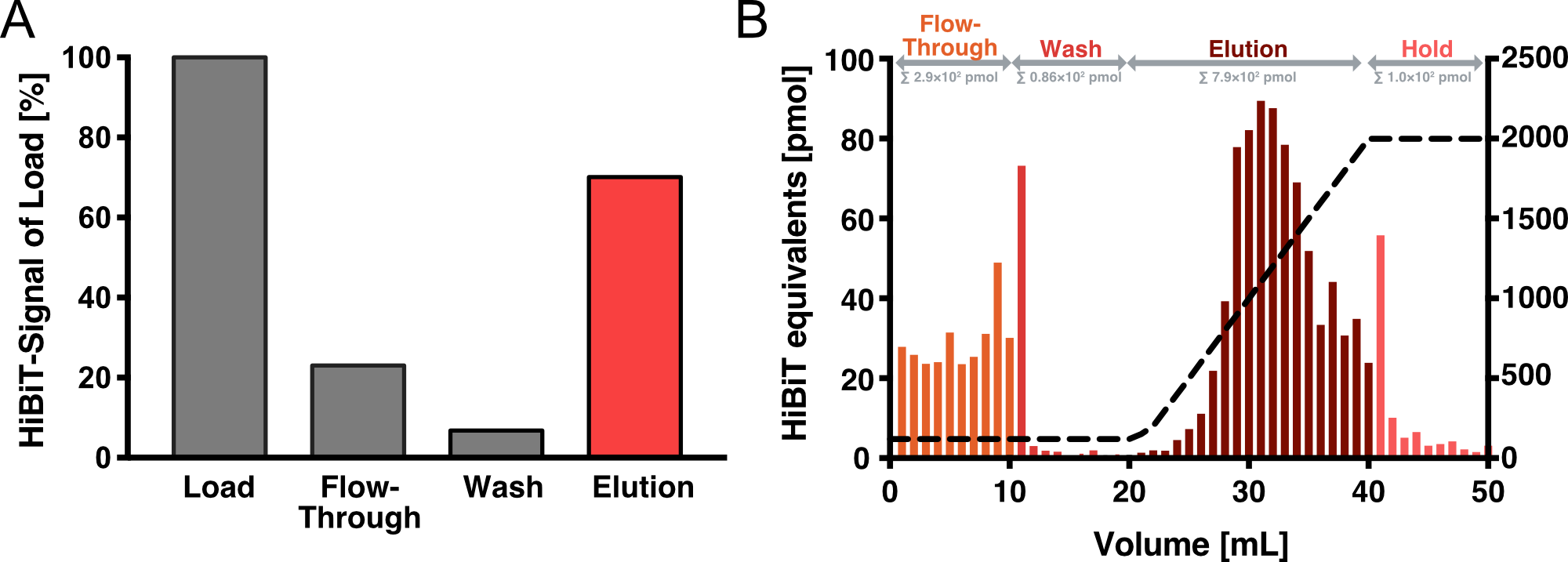
Affinity Purification
We have established Heparin affinity chromatography as our method of choice for VLP purification. Figure 4 shows an exemplary chromatogram of a purification run with an increasing NaCl gradient for elution. Fractions of 1 ml were collected for each NaCl concentration up to 2 M and analyzed with an HiBiT assay to determine the absolute amount of Gag. After elution, we collected another 10 fractions with 2 M NaCl. As can be seen, these fractions showed almost no HiBiT signal, indicating that all vesicles were eluted from the column. In total, approximately 4 % of the HiBiT signal was detected in the flow-through, 5% in the wash and 55% in the elution fraction.
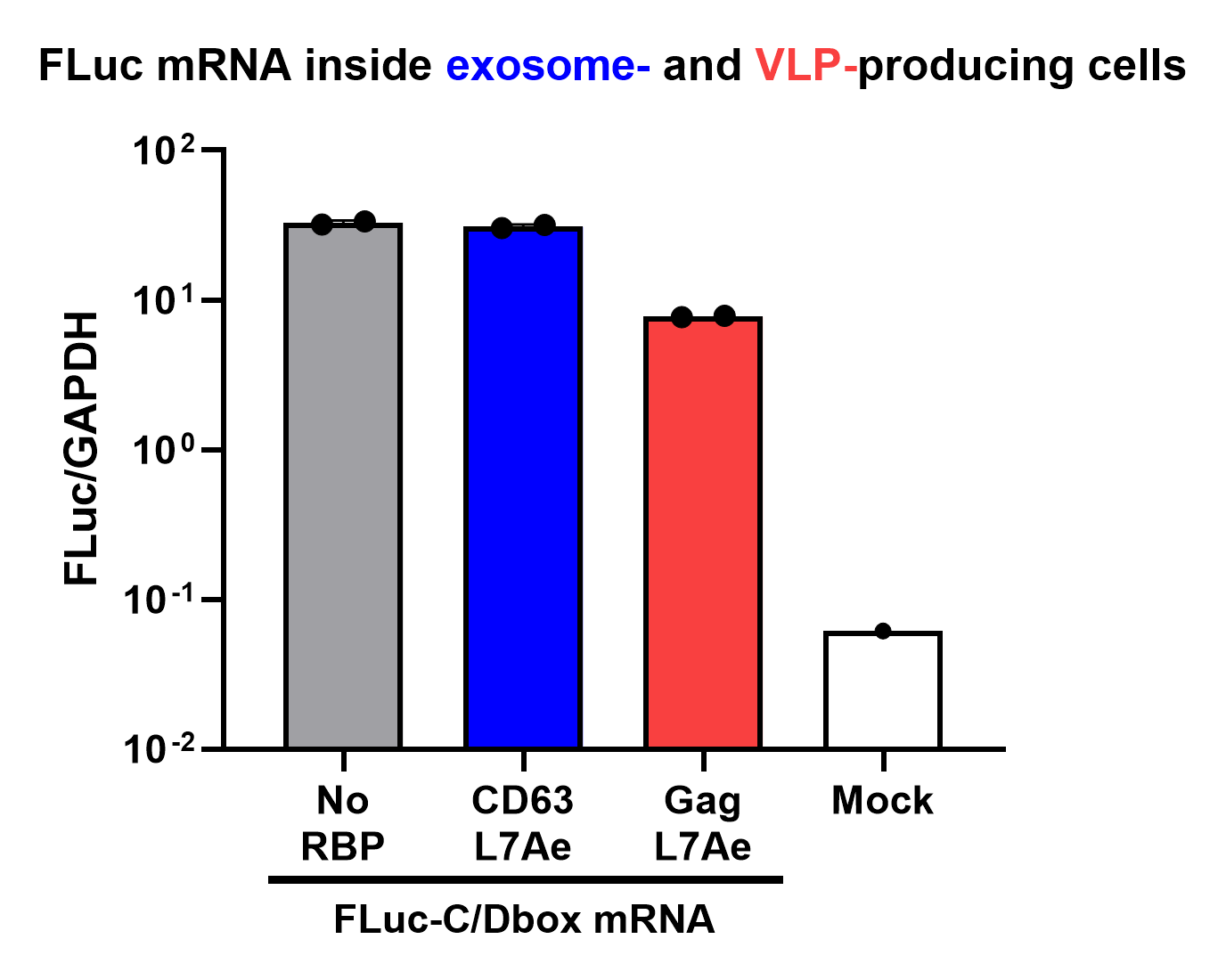
qPCR Analysis
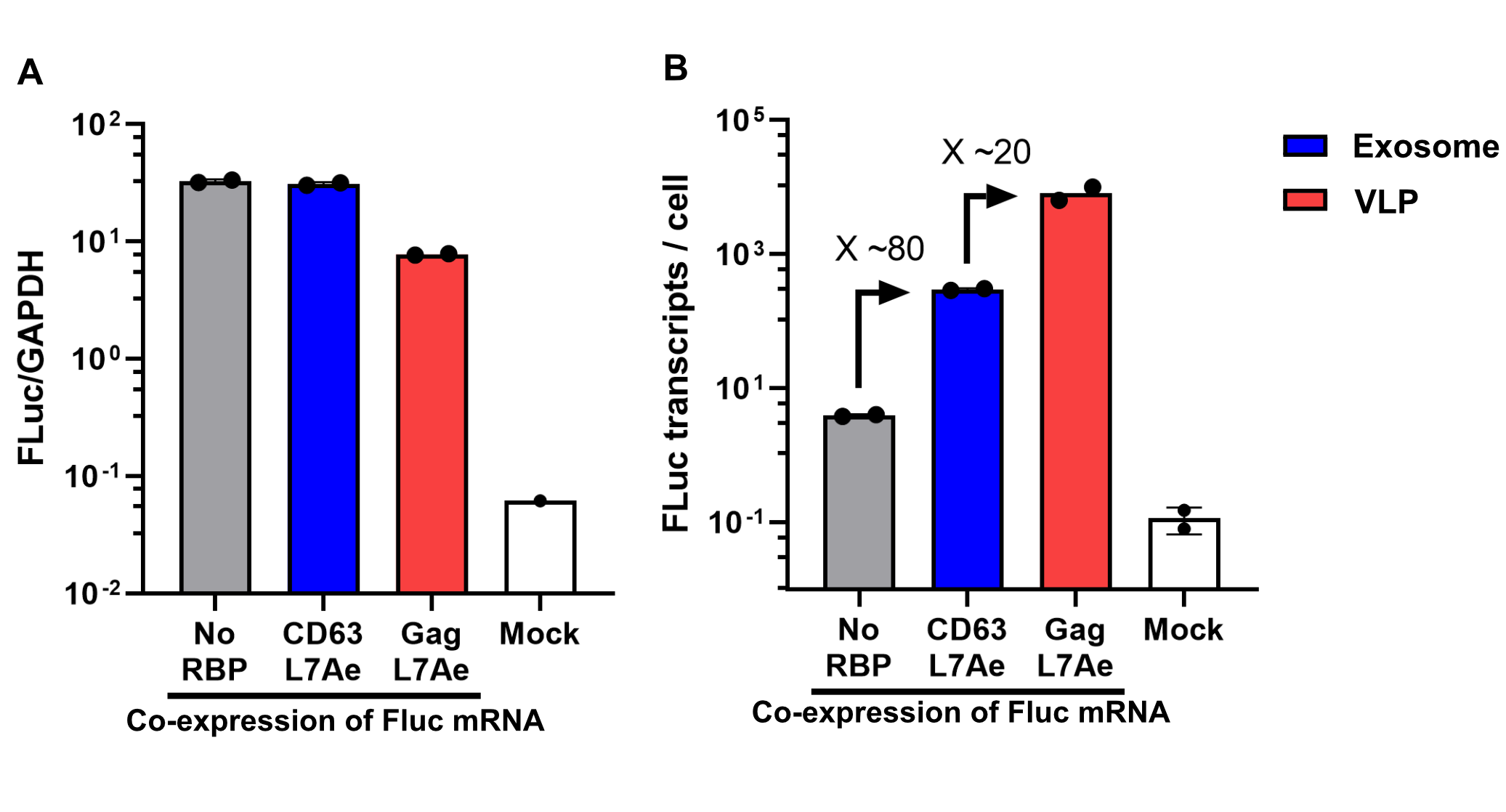
Finally, the BioBrick Part we designed works as expected since chosen RNA information is successfully transported into VLPs through L7Ae-C/D box interaction. This has been proven through qPCR analysis of VLP content. FLuc mRNA is calculated with delta Ct analysis, where GAPDH housekeeping gene is used as background. Also taking cell confluency (70-80%) into account, we can calculate the accurate amount of Fluc RNA content/cell. On the graph below you can observe a 100-fold increase in FLuc signal/cell with our validated VLP BioBrick Part, compared to control. Furthermore, this part performs 20-fold better than a parallel construct used for exosomes.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 3019
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 2319
References
- ↑ HIV-1 Gag: a Molecular Machine Driving Viral Particle Assembly and Release Heinrich G. Göttlinger Department of Cancer Immunology and AIDS, Dana-Farber Cancer Institute, and Department of Pathology, Harvard Medical School, Boston
- ↑ https://www.promega.de/products/protein-quantitation-and-detection/protein-quantitation/nano-glo-hibit-lytic-detection-system/?catNum=N3030
- ↑ Gagnon, Keith T et al. “Signature amino acids enable the archaeal L7Ae box C/D RNP core protein to recognize and bind the K-loop RNA motif.” RNA (New York, N.Y.) vol. 16,1 (2010): 79-90. doi:10.1261/rna.1692310
- ↑ Lilley, D. M. J. (2019). "The L7Ae proteins mediate a widespread and highly functional protein–RNA interaction." The Biochemist 41(2): 40-44.
