Difference between revisions of "Part:BBa K3113100"
Theresakeil (Talk | contribs) |
Theresakeil (Talk | contribs) |
||
| Line 48: | Line 48: | ||
<html> | <html> | ||
<figure class="figure"> | <figure class="figure"> | ||
| − | <img src="" width=" | + | <img src="https://2019.igem.org/wiki/images/c/cc/T--Munich--Validation_Purification.png" width="460" height="360" class="figure-img img-fluid rounded" alt="VLP Purification."> |
<figcaption class="figure-caption"><b>Figure 3: Heparin chromatography purification of VLPs.</b> Gag vesicles were purified from cell culture medium on Heparin columns. n = 1 biological replicates | <figcaption class="figure-caption"><b>Figure 3: Heparin chromatography purification of VLPs.</b> Gag vesicles were purified from cell culture medium on Heparin columns. n = 1 biological replicates | ||
| Line 61: | Line 61: | ||
<html> | <html> | ||
<figure class="figure"> | <figure class="figure"> | ||
| − | <img src="" width="400" height="360" class="figure-img img-fluid rounded" alt="HEK293T: unloaded vs. loaded VLPs."> | + | <img src=" https://2019.igem.org/wiki/images/1/1e/T--Munich--VLP_DLS_HEK_empty_vs_loaded.png" width="400" height="360" class="figure-img img-fluid rounded" alt="HEK293T: unloaded vs. loaded VLPs."> |
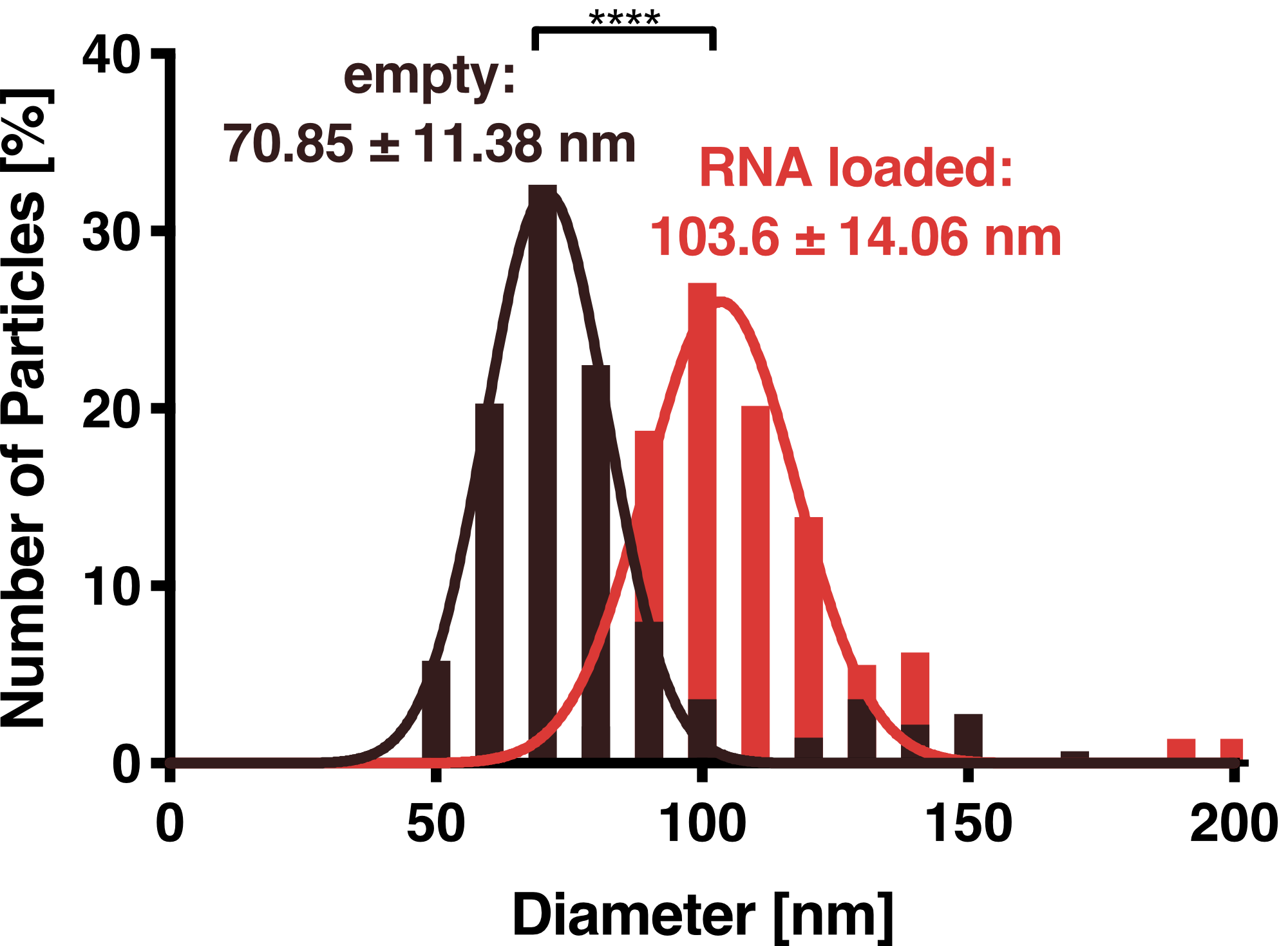
<figcaption class="figure-caption"><b>Figure 4: Dynamic light scattering (DLS) measurement of VLPs.</b> Purified VLPs either with or without the adapter protein L7Ae were measured. Unloaded and loaded vesicles showed a narrow size distribution with mean diameters of 71 and 104 nm, respectively. | <figcaption class="figure-caption"><b>Figure 4: Dynamic light scattering (DLS) measurement of VLPs.</b> Purified VLPs either with or without the adapter protein L7Ae were measured. Unloaded and loaded vesicles showed a narrow size distribution with mean diameters of 71 and 104 nm, respectively. | ||
| Line 75: | Line 75: | ||
<html> | <html> | ||
<figure class="figure"> | <figure class="figure"> | ||
| − | <img src="" width="400" height="360" class="figure-img img-fluid rounded" alt="Size distribution frequency of VLPs from HEK293T."> | + | <img src=" https://2019.igem.org/wiki/images/a/a4/T--Munich--TEM_VLP_HEK_loaded_vs_unlaoded.png" width="400" height="360" class="figure-img img-fluid rounded" alt="Size distribution frequency of VLPs from HEK293T."> |
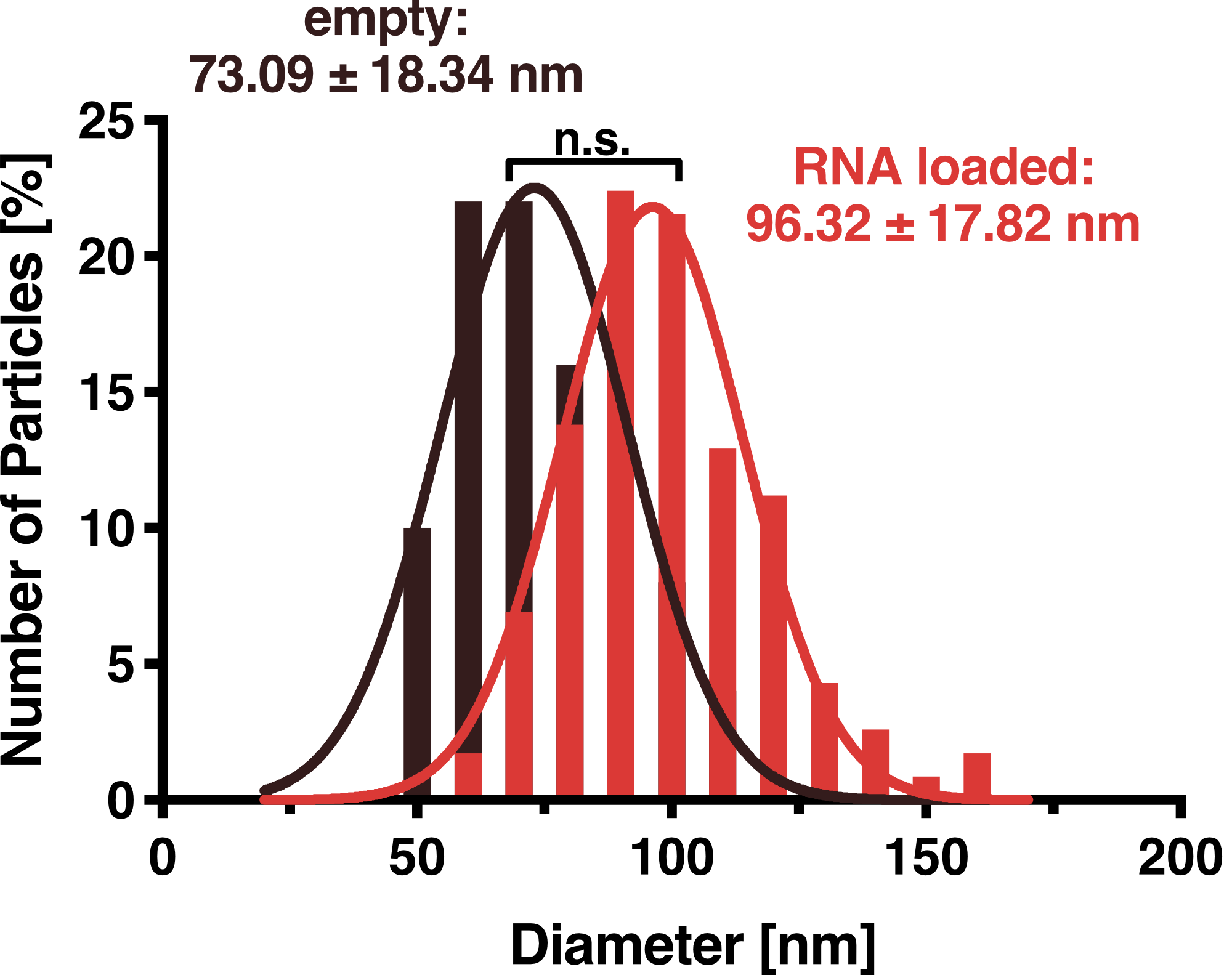
<figcaption class="figure-caption"><b>Figure 5:</b> This Figure shows an analysis of particle diameter distribution for VLPs secreted from HEK293T cells calculated from 50 (unloaded, red) and 116 (loaded, blue) particles, respectively. The inset shows exemplary particles as seen in TEM highlighted with dashed lines (scalebar 100 nm). A Gaussian fit was performed on the data and the average diameter was calculated as 72 nm and 96 nm for unloaded and loaded VLPs, respectively. | <figcaption class="figure-caption"><b>Figure 5:</b> This Figure shows an analysis of particle diameter distribution for VLPs secreted from HEK293T cells calculated from 50 (unloaded, red) and 116 (loaded, blue) particles, respectively. The inset shows exemplary particles as seen in TEM highlighted with dashed lines (scalebar 100 nm). A Gaussian fit was performed on the data and the average diameter was calculated as 72 nm and 96 nm for unloaded and loaded VLPs, respectively. | ||
Revision as of 22:22, 21 October 2019
Gag HIV-1
This sequence codes for the coat protein of the human immunodeficiency virus, which mediates the essential events in virion assembly, including binding the plasma membrane, making the protein-protein interactions necessary to create spherical particles.
Usage
For ALiVE, two vesicles were tested. One kind of vesicle is based on the Gag-p24 capsid protein of HIV. We used this protein fused together with RNA binding proteins to build and load virus-like vesicles for non-invasive cell monitoring.
Biologie
Gag-p24 is the group-specific antigen from the lentivirus HIV. It assembles at the plasma membrane and leads to the budding of vesicles.[1]
Characterization
Protein Structure

HiBiT
To furtherly prove the BioBrick Part we designed works as expected we performed a HiBiT split luciferase assay, which shows luminescent signal detected in formed VLPs. The following graph shows the amount of expressed Gag-HiBiT in cells compared to the amount measured in the supernatant. With this assay, we could show that 45 percent of Gag is exported to the supernatant in the form of vesicles.

Purification
Contrary to the exosomal protein CD63, the structural Gag protein forming the VLP shell contains no transmembrane domains and binds the vesicle membrane only on the cytoplasmic side. Therefore, it lacks extracellular regions that could be used to insert an affinity tag. We thus tested Heparin affinity chromatography, which has been reported previously (Reiter et al 2019) as a suitable method for Gag-based vesicle purification. As Figure 5 shows, we were able to robustly purify VLPs using Heparin chromatography. Overall, we could recover more than 50 % of VLPs with this purification method.

Dynamic Light Scattering
DLS measurements of VLP particles showed a narrow Gaussian size distribution indicating that the samples are very homogenous. Interestingly, a shift of about 30 nm is seen between cargo-loaded and unloaded VLPs; cargo-loaded particles have a mean diameter of 104 ± 14 nm, whereas unloaded vesicles showed a mean diameter of 71 ± 11 nm.
Transmission Electron Microscopy
To further validate that our vesicles are intact and properly shaped, we prepared purified VLPs and exosomes for transmission electron microscopy (TEM). This microscopy technique is based on a high-energy beam of electrons shown through a very thin sample fixed on a grid and allows high-resolution imaging.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1307
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 607
References
- ↑ HIV-1 Gag: a Molecular Machine Driving Viral Particle Assembly and Release Heinrich G. Göttlinger Department of Cancer Immunology and AIDS, Dana-Farber Cancer Institute, and Department of Pathology, Harvard Medical School, Boston
