Difference between revisions of "Part:BBa K3052001"
| Line 35: | Line 35: | ||
===Characterization=== | ===Characterization=== | ||
| − | Ptrc and CS genes was obtained by PCR flanked two Type IIS sites BsaI and was used for Golden Gate Assembly to construct the expression vectors. The length of limonene synthase | + | Ptrc and CS genes was obtained by PCR flanked two Type IIS sites BsaI and was used for Golden Gate Assembly to construct the expression vectors. The length of limonene synthase CS is 1632 bp. Here is the gel results confirming success amplification. |
<div>[[File:T--XJTU-CHINA--CS-PCR.png |700px|thumb|center|<b>Figure 2:</b> Gel results of PCR amplification of CS ]]</div> | <div>[[File:T--XJTU-CHINA--CS-PCR.png |700px|thumb|center|<b>Figure 2:</b> Gel results of PCR amplification of CS ]]</div> | ||
| − | The expression of limonene synthase and linalool synthases are under the regulation of Ptrc. RT-qPCR and SDS-PAGE were conducted to test their expressions, as well as Gas Chromatography analysis was used to detect limonene | + | The expression of limonene synthase and linalool synthases are under the regulation of Ptrc. RT-qPCR and SDS-PAGE were conducted to test their expressions, as well as Gas Chromatography analysis was used to detect limonene yields. Click to see our protocol. |
| − | We measured the transcription level of CS and LSs of four different strains, E.coli harboring only pGPP vector, E.coli with dual | + | We measured the transcription level of CS and LSs of four different strains, E.coli harboring only pGPP vector, E.coli with dual plasmids pGPP+pDHJ (Stain B in Fig 3), pGPP+pLGF (Stain C in Fig 3), pGPP+pCS (Stain A in Fig 3),and relative mRNA levels compared with the blank strain only harboring pGPP are shown in Fig 4. |
<div>[[File:T--XJTU-CHINA--DM7.png |700px|thumb|center|<b>Figure 3:</b>RT-qPCR result of linalool synthase DHJ, LGF and limonene synthase CS]]</div> | <div>[[File:T--XJTU-CHINA--DM7.png |700px|thumb|center|<b>Figure 3:</b>RT-qPCR result of linalool synthase DHJ, LGF and limonene synthase CS]]</div> | ||
| − | As shown in Fig 4, compared to blank strain, the relative mRNA level of three strains were much higher | + | As shown in Fig 4, compared to blank strain, the relative mRNA level of three strains were much higher, and clearly showed the efficient transcription and expression of CS gene in engineered E.coli. |
| − | We also conducted SDS-PAGE of whole cell proteins to ensure | + | We also conducted SDS-PAGE of whole cell proteins to ensure its expression. As shown in Fig 6, compared to blank strain only with pGPP vector, CS protein was expressed but yields were not high, and because of the leaky expression, expression level with IPTG induction were just a little bit higher than the level without IPTG induction. |
<div>[[File:T--XJTU-CHINA--CS-PAGE.png |700px|thumb|center|<b>Figure 4:</b>SDS-PAGE result of CS: Control group: sample from E.coli transformed with pGPP; Experimental group: sample from E.coli transformed with pGPP and pCS (+:with IPTG induction/-: without IPTG induction)]]</div> | <div>[[File:T--XJTU-CHINA--CS-PAGE.png |700px|thumb|center|<b>Figure 4:</b>SDS-PAGE result of CS: Control group: sample from E.coli transformed with pGPP; Experimental group: sample from E.coli transformed with pGPP and pCS (+:with IPTG induction/-: without IPTG induction)]]</div> | ||
| Line 59: | Line 59: | ||
<div>[[File:T--XJTU-CHINA--CSGC2.png |700px|thumb|center|<b>Figure 6:</b>Gas Chromatography result of standard sample. (A1)The sample of 1ul limonene is soluble in 1ml n-hexadecane. (A2) Magnified view of peak at 1.37 min of retention time in A1. (B1) The sample of 0.3ul limonene was soluble in 1ml n-hexadecane. (B2) Magnified view of peak at 1.37 min of retention time in B1. (C1) Samples from E.coli (transformed with pGPP and pCS) incubated in YT culture medium for 48h. (C2) Magnified view of peak at 1.37 min of retention time in C1. ]]</div> | <div>[[File:T--XJTU-CHINA--CSGC2.png |700px|thumb|center|<b>Figure 6:</b>Gas Chromatography result of standard sample. (A1)The sample of 1ul limonene is soluble in 1ml n-hexadecane. (A2) Magnified view of peak at 1.37 min of retention time in A1. (B1) The sample of 0.3ul limonene was soluble in 1ml n-hexadecane. (B2) Magnified view of peak at 1.37 min of retention time in B1. (C1) Samples from E.coli (transformed with pGPP and pCS) incubated in YT culture medium for 48h. (C2) Magnified view of peak at 1.37 min of retention time in C1. ]]</div> | ||
| − | The results show that the retention time of limonene is more clear at 1.37 minutes after optimization when the amount of limonene is small. It shares the same retention time our experimental result of samples from E.coli that is transformed with pGPP and pCS | + | The results show that the retention time of limonene is more clear at 1.37 minutes after optimization when the amount of limonene is small. It shares the same retention time with our experimental result of samples from E.coli that is transformed with pGPP and pCS. And the important GC-MS result showed the the MS profile is exactly the same with compound library and literature, further confidently confirming its production(See Fig below). This result indicates that GPP production can be achieved by precursor circuit, and most importantly, our limonene synthase can work properly to convert GPP to limonene even though the yield of limonene remains low, which needs to be further improved by more metabolic engineering of the strain. |
<div>[[File:T--XJTU-CHINA--CSGC3.png |700px|thumb|center|<b>Figure 7:</b>Comparison of limonene production of E.coli strain harboring CS gene with limonene standard, confirmed its production.]]</div> | <div>[[File:T--XJTU-CHINA--CSGC3.png |700px|thumb|center|<b>Figure 7:</b>Comparison of limonene production of E.coli strain harboring CS gene with limonene standard, confirmed its production.]]</div> | ||
| Line 66: | Line 66: | ||
<div>[[File:T--XJTU-CHINA--GCMS-B.png |700px|thumb|center|<b>Figure 8:</b> GC-MS result of limonene production by E.coli strain harboring CS gene: (A)The gas chromatography retention time of 9.8 minutes and (B) the mass spectrometry base peak at 93m/z (Figure B) are characteristic of limonene. (A) We analysed a positive limonene-containing control that yielded a strong peak as indicated by the black line as well as a negative control where no GPP was added to the assay, which yielded no peak as indicated by the red line. In comparison, our expressed limonene synthase that was incubated with GPP produced by pGPP (indicated by the blue line) shows the indicative limonene peak at 9.8 minutes. (B) Cross reference with a compound library revealed that the limonene synthase sample's peak chromatography at 9.8 minutes (top panel) matches the library's d-limonene peak chromatography (bottom panel).]]</div> | <div>[[File:T--XJTU-CHINA--GCMS-B.png |700px|thumb|center|<b>Figure 8:</b> GC-MS result of limonene production by E.coli strain harboring CS gene: (A)The gas chromatography retention time of 9.8 minutes and (B) the mass spectrometry base peak at 93m/z (Figure B) are characteristic of limonene. (A) We analysed a positive limonene-containing control that yielded a strong peak as indicated by the black line as well as a negative control where no GPP was added to the assay, which yielded no peak as indicated by the red line. In comparison, our expressed limonene synthase that was incubated with GPP produced by pGPP (indicated by the blue line) shows the indicative limonene peak at 9.8 minutes. (B) Cross reference with a compound library revealed that the limonene synthase sample's peak chromatography at 9.8 minutes (top panel) matches the library's d-limonene peak chromatography (bottom panel).]]</div> | ||
| − | The results indicate that the | + | The results indicate that the MS profile is exactly the same with compound library and literature,confirming its production. |
Revision as of 21:59, 21 October 2019
(4S)-limonene synthase
The limonene synthase (CS) sequence used throughout our project which converts GPP to limonene is an E. coli codon-optimized version of a truncated sequence from M. spicata previously described(Hyatt et al., 2007).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Characterization
Background
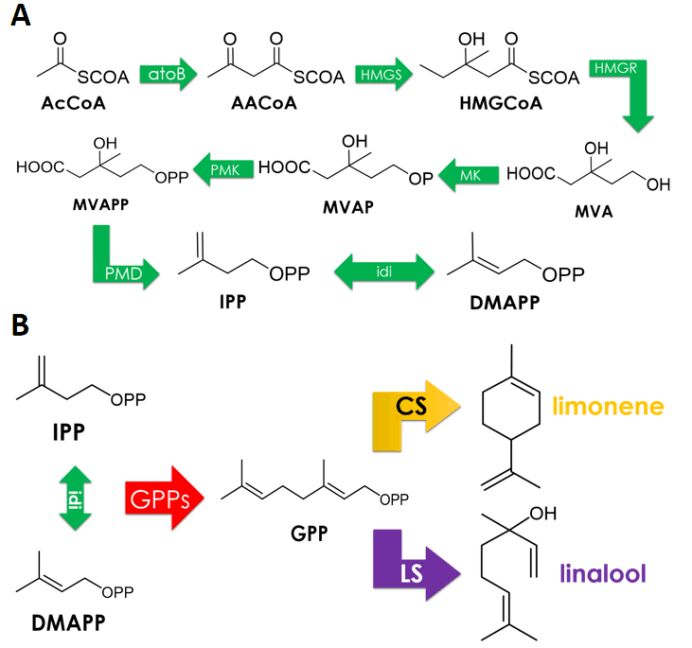
In our study, we aim to achieve limonene and linalool synthesis in E.coli DH5α. According to 2018 GreatBay_China team’s experience, no target product was detected using gas chromatography when carrying out shake-flask fermentation with this strain induced by 25uM IPTG for 24 hours due to the lack of endogenous MVA pathways wherein GPP is produced. Thus we decided to co-express an MVA pathway. 2018 GreatBay_China generously gave us one plasmid pJBEI6409, which contains a MVA pathway in addition to an GPPs-CS operon. We reconstructed this plasmid and get a plasmid only contains a MVA pathway.
We have used MVA synthesis pathway which is common in plants. Since limonene and linalool have the same synthetic precursor GPP, we have divided the synthesis pathway into two parts:
1. (BBa_K3052001) IPTG inducible precursor circuit which contains eight enzymes of MVA pathway enable conversion from AcCoA to GPP: atoB, HMGS, HMGR, MK, PMK, PMD, idi, trGPPs.
2. (BBa_K3052004, BBa_K3052010) two Production Circuits: limonene synthase or linalool synthase regulated by a ptrc promoter in E. coli.
Characterization
Ptrc and CS genes was obtained by PCR flanked two Type IIS sites BsaI and was used for Golden Gate Assembly to construct the expression vectors. The length of limonene synthase CS is 1632 bp. Here is the gel results confirming success amplification.
The expression of limonene synthase and linalool synthases are under the regulation of Ptrc. RT-qPCR and SDS-PAGE were conducted to test their expressions, as well as Gas Chromatography analysis was used to detect limonene yields. Click to see our protocol. We measured the transcription level of CS and LSs of four different strains, E.coli harboring only pGPP vector, E.coli with dual plasmids pGPP+pDHJ (Stain B in Fig 3), pGPP+pLGF (Stain C in Fig 3), pGPP+pCS (Stain A in Fig 3),and relative mRNA levels compared with the blank strain only harboring pGPP are shown in Fig 4.
As shown in Fig 4, compared to blank strain, the relative mRNA level of three strains were much higher, and clearly showed the efficient transcription and expression of CS gene in engineered E.coli. We also conducted SDS-PAGE of whole cell proteins to ensure its expression. As shown in Fig 6, compared to blank strain only with pGPP vector, CS protein was expressed but yields were not high, and because of the leaky expression, expression level with IPTG induction were just a little bit higher than the level without IPTG induction.
Both the results of above RT-qPCR and SDS-PAGE demonstrate the expression of the limonene synthase CS.
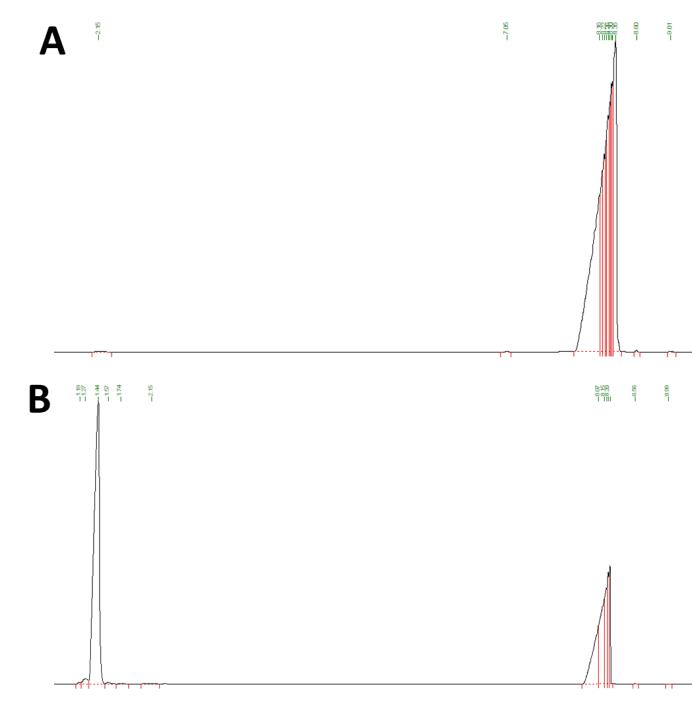
GC analysis will confirm whether the expressed protein is functional. Firstly we analyzed a positive limonene-containing control that yielded a strong peak as indicated by the black line with the retention time abound 1.44 minutes. The results show that there is no overlap of retention time between n-Hexadecane and limonene, and indicates the GC method is effective for testing. In comparison, we expressed the limonene generator with trc promoter in DH5α E. coli and lysed the cell culture, extracted with n- hexadecane, and the extraction was subjected to gas chromatography mass spectrometry.

The results show that the retention time of limonene is more clear at 1.37 minutes after optimization when the amount of limonene is small. It shares the same retention time with our experimental result of samples from E.coli that is transformed with pGPP and pCS. And the important GC-MS result showed the the MS profile is exactly the same with compound library and literature, further confidently confirming its production(See Fig below). This result indicates that GPP production can be achieved by precursor circuit, and most importantly, our limonene synthase can work properly to convert GPP to limonene even though the yield of limonene remains low, which needs to be further improved by more metabolic engineering of the strain.

The results indicate that the MS profile is exactly the same with compound library and literature,confirming its production.