Difference between revisions of "Part:BBa K784001"
Konstantinos (Talk | contribs) |
Konstantinos (Talk | contribs) (→Expression of CDO in OnePot PURE) |
||
| Line 10: | Line 10: | ||
=='''Expression of CDO in OnePot PURE'''== | =='''Expression of CDO in OnePot PURE'''== | ||
| − | [[File:T--EPFL--Detection_cdo_result.jpg|thumb|600px|left|<b>Figure 3.</b> Fluorescein standard curve used to quantify fluorescent intensity. Slight discrepancy from the linear trend at high concentration is most likely due to oversaturation of signal. | + | [[File:T--EPFL--Detection_cdo_result.jpg|thumb|600px|left|<b>Figure 42</b> Fluorescent intensity of BL21 expressing GFP under T7 (blue), and PR4 (Green). The T7 promoter lead to about 2.6 as much protein being produced over a 16 hour timescale. Fluorescent intensity was determined using the fluorescein standard and logarithmic curves. |
| + | ]] | ||
| + | [[File:T--EPFL--Detection_cdo_result.jpg|<b>Figure 3.</b> Fluorescein standard curve used to quantify fluorescent intensity. Slight discrepancy from the linear trend at high concentration is most likely due to oversaturation of signal. | ||
]] | ]] | ||
<br/> | <br/> | ||
Revision as of 16:24, 21 October 2019
pT7_RBS_xyIE reporter gene
This part's purpose is to testify to the presence of the T7 RNA Polymerase by the xyIE reporter gene from iGEM2006_Edinburgh. It consists of an RNA T7 promoter, an RBS, a reporter gene- xyIE, and a T7 terminator. The assay for xyIE is very simple, refer to the xyIE basic part for instructions.
The xyIE+RBS part is between XhoI and XmaI restriction sites, therefore can be excised by these enzymes.
Expression of CDO in OnePot PURE
Figure 3. Fluorescein standard curve used to quantify fluorescent intensity. Slight discrepancy from the linear trend at high concentration is most likely due to oversaturation of signal.
Group: EPFL, 2019
Authors: Laura Kvedarauskaite, Konstantinos Ragios
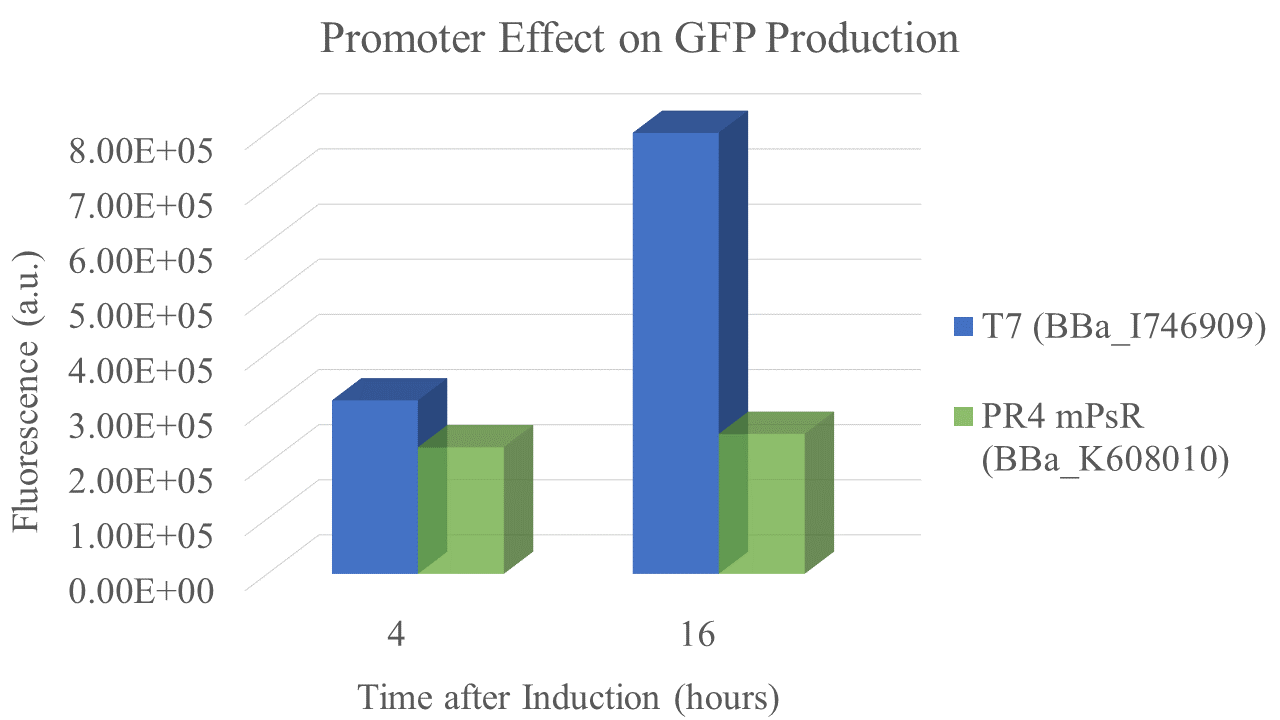
Summary: We compared the amount of GFP expressed under a constitutive promoter (medium promoter, strong RBS) to T7 expression. Some proteins fold better under constitutive promoters; however, nobody had yet directly compared the amount of protein produced between constitutive vs. T7 expression.
Methods
BioBricks were transformed and expressed in E. coli (BL21). BL21 cells were cultured to an OD600=0.6 and 100 uL of culture was transferred into a 96 well plate. Colonies were transfered in quadruplicate. The fluorescence intensity of GFP was measured with a multi-mode microplate reader. The iGEM standardized fluorescence protocol was used for fluorescence measurement standardization (https://www.protocols.io/view/calibration-protocol-plate-reader-fluorescence-cal-6zrhf56).
Results
We found that the T7 promoter produced about 2.6 times as much fluorescent signal as the constitutive PR4 promoter, indicating that T7 is much more efficient at producing GFP (Fig. 5). Interestingly, the production of GFP under PR4 did not increase beyond the level observed at 4 hours after inoculation. It seems that PR4 leads to an initial production of protein; however, after the initial expression the promoter seems to be shut off.
Promoter and RBS:
PR4: medium Promoter (J23110) strong RBS (B0034)
T7: T7 Promoter (BBa_I746909)
| sample | PR4 | T7 |
| Fluorescence 4 hrs (a.u.) | 2.24E+05 | 3.09E+05 |
| Fluorescence 16 hrs (a.u.) | 2.48E+05 | 8.00E+05 |
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 368
Illegal NgoMIV site found at 540
Illegal AgeI site found at 891 - 1000COMPATIBLE WITH RFC[1000]