Difference between revisions of "Part:BBa K3037000"
(→Overview: listing) |
(→Overview) |
||
| Line 25: | Line 25: | ||
* LacO promoter and LacI inhibitor | * LacO promoter and LacI inhibitor | ||
* Kanamycin resistance | * Kanamycin resistance | ||
| − | * Needs T7 RNA polymerase | + | * Needs T7 RNA polymerase (a viral RNA polymerase for high expression) |
| − | ** | + | **For optimal results use K3037000 in combination with a pRARE plasmid, carried by the used <span style="font-style: italic;">Escherichia coli (E. coli)</span> strain |
| − | **In case you use pRARE (Cm), you need to grow in a media | + | **In case you use pRARE (Cm), you need to grow in a media containing both antibiotics - Kanamycin (Kan) and Chloramphenicol (Cm) |
| − | ** | + | **Alternatively use a strain which can express T7 from their genome, regular <span style="font-style: italic;">E. coli</span> strains do not express T7! |
* Contains SP6 site which is a commonly used primer site for sequencing the inserted part | * Contains SP6 site which is a commonly used primer site for sequencing the inserted part | ||
* Codon optimized for <span style="font-style: italic;">E. coli.</span> | * Codon optimized for <span style="font-style: italic;">E. coli.</span> | ||
| − | * Very well established expression plasmid for recombinant proteins | + | * Very well established expression plasmid for recombinant proteins in <span style="font-style: italic;">E. coli.</span>! From ligation to expression in just 24 hours! |
=== Biology === | === Biology === | ||
Revision as of 13:12, 21 October 2019
pOCC97 plasmid backbone for expression (optimized)
| pOCC97 | |
|---|---|
| Function | Expression |
| Use in | Escherichia coli pRARE T7 |
| RFC standard | RFC 10 |
| Submitted by | Team: TU_Dresden 2019 |
Contents
- 1 Overview
- 2 Characterization
- 2.1 Outline
- 2.2 Experiments in Detail
- 2.2.1 1) Expression of different proteins, growing curve (BBa_3037003)(BBa_K3037007)
- 2.2.2 2) Expression of proteins with the plasmid before and after optimization at different temperatures
- 2.2.3 3) SDS-PAGEs for the expression assay over the time of Full Construct (BBa_K3037003)
- 2.2.4 4) Image analysis of the expression in the SDS-PAGEs with imageJ
- 2.2.5 5) Expression of different BioBricks in pOCC97
- 3 Sequence
- 4 Design Notes
- 5 References
Overview
The TU Dresden 2019 team designed this BioBrick in order to express its fusion protein (more information).
Features:
- IPTG inducible
- LacO promoter and LacI inhibitor
- Kanamycin resistance
- Needs T7 RNA polymerase (a viral RNA polymerase for high expression)
- For optimal results use K3037000 in combination with a pRARE plasmid, carried by the used Escherichia coli (E. coli) strain
- In case you use pRARE (Cm), you need to grow in a media containing both antibiotics - Kanamycin (Kan) and Chloramphenicol (Cm)
- Alternatively use a strain which can express T7 from their genome, regular E. coli strains do not express T7!
- Contains SP6 site which is a commonly used primer site for sequencing the inserted part
- Codon optimized for E. coli.
- Very well established expression plasmid for recombinant proteins in E. coli.! From ligation to expression in just 24 hours!
Biology
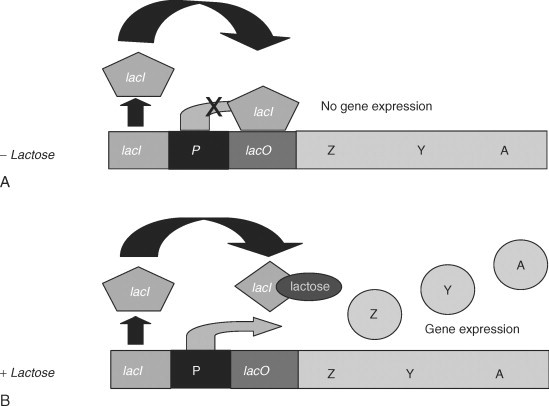
The lactose operon (lac operon) is a polycistronic bacterial operon that encodes the genes of lactose metabolism. It is consists of three structural genes: a promoter, an operator, a terminator. A bacterial cell synthesizes enzymes involved in lactose metabolism only under the two conditions: in the presence of lactose and when the cells lack glucose. [1]
The regulation of the lac operon occurs according to the principle of negative feedback: the more lactose is present in the environment - the more enzymes for its catabolism is synthesized (positive direct connection); the more enzymes are present in a cell - the less lactose remains, and finally, the less lactose in the enironment - the less enzymes are produced (double negative feedback).[2]
In the absence, or at a low concentration of lactose in the cell, the repressor protein, which is a product of the LacI monocistronic operon, reversibly binds to the operator region and inhibits transcription. Thus, in the absence of lactose in the cell, enzymes for lactose metabolism are not synthesized. Besides, if the glucose concentration in the cell is sufficient to maintain metabolism, activation of the lactose operon also does not occur. The promoter sequence of the lactose operon is weak, therefore, even in the absence of a repressor protein in the operator site, transcription is practically not initiated.
When the concentration of glucose in the cell decreases, the enzyme adenylate cyclase activates, which catalyzes the conversion of ATP to the cyclic form - cAMP (the cyclic form of AMP). Glucose is an inhibitor of the enzyme adenylate cyclase and activates phosphodiesterase, an enzyme that catalyzes the conversion of the cAMP molecule to AMP. cAMP binds to a catabolism activating protein (CAP), and a complex is formed that interacts with the promoter of the lactose operon, changes its conformation, and increases the affinity of RNA polymerase for this site. In the presence of lactose, expression of the operon genes occurs.[2]
Characterization
Outline
1) Expression of different proteins, growing curve (BBa_3037003)(BBa_K3037007)
2) Expression of proteins with the plasmid before and after optimization at different temperatures
3) SDS-PAGEs for the expression assay over the time of Full Construct (BBa_K3037003)
4) Image analysis of the expression in the SDS-PAGEs with imageJ
5) Expression of different BioBricks in pOCC97
Experiments in Detail
1) Expression of different proteins, growing curve (BBa_3037003)(BBa_K3037007)
Two different proteins were expressed using this backbone: HRP (BBa_K3037007) and a bigger fusion protein (BBa_K3037003).
The organism used for the expression was E. coli pRARE T7
The results prove that when expressed, non of the two proteins affect the growing curve of the bacteria. Also, it can be seen how the growing ratio is lower in the culture that is expressing a bigger protein.
Growing curve (When MBP-HRP (BBa_K3037008) composite BioBrick is expressed)
2) Expression of proteins with the plasmid before and after optimization at different temperatures
To use this backbone as an expression vector adapted to the iGEM standards, we had to mutate it first to include the Prefox and Suffix of the BioBrick Assembly. Before to do that, we thought that it will be great to have a comparison between the expression before and after the mutation, a process that we called optimization.
We found that the original plasmid had a XbaI sequence that we used to insert our BioBrick expressed protein BBa_K3037003. This ilegal restriction site was removed during the optimization process. The XbaI restriction site was placed after the promoter (T7) and before the RBS sequence of the plasmid. However, since we were using the RFC 25 standard for fusion proteins, the inserted protein contained this was its own RBS (included in the prefix of the RFC 25 standard).
Then, for the optimization we removed the XbaI restriction site and included a Prefix and Suffix of the RFC 10 standard after the RBS of the plasmid. After that we used the BioBrick assembly method to insert there our protein BBa_K3037003, which has its own RBS due to the RFC 25 standard.
The comparison of the growing curve shows that, although, the new plasmid adapted to the RFC 10 standard didn't affect the growing rate of the bacteria, as the original one, but the expression of the protein was improved.
3) SDS-PAGEs for the expression assay over the time of Full Construct (BBa_K3037003)
Comparison of the expression of MBP-HRP (BBa_K3037008) and Full Construct (BBa_3037003)
Expression of full construct in pOCC97 not optimized at 18ºC
Expression of Full Construct in pOCC97 at 37ºC
Expression of Full Construct in pOCC97 optimized
4) Image analysis of the expression in the SDS-PAGEs with imageJ
Different induction concentration not optimized at 18ºC and at 37ºC
Not optimized at different temperature at 0.2 mM, 0.5 mM and 1 mM IPTG expression induction
Different temperature optimized 1mM
Different induction concentration optimizedat 18 degrees
Comparison optimized vs not optimized
Conclusion
Based on this analysis can be concluded that optimal conditions for the expression of BBa_3037003 are 18ºC and 0.5 mM IPTG. Also the optimization process is better than the not optimazed as the expression is more stable over time
5) Expression of different BioBricks in pOCC97
With this experiment we proved that is easy to express diferent and functional BioBricks with this vector
Sequence
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 5283
Illegal SpeI site found at 2
Illegal PstI site found at 16
Illegal NotI site found at 9
Illegal NotI site found at 5289 - 21INCOMPATIBLE WITH RFC[21]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 5283
Illegal BglII site found at 5169 - 23INCOMPATIBLE WITH RFC[23]Illegal prefix found at 5283
Illegal suffix found at 2 - 25INCOMPATIBLE WITH RFC[25]Illegal prefix found at 5283
Plasmid lacks a suffix.
Illegal XbaI site found at 5298
Illegal SpeI site found at 2
Illegal PstI site found at 16
Illegal NgoMIV site found at 342
Illegal NgoMIV site found at 3389
Illegal NgoMIV site found at 3549
Illegal NgoMIV site found at 5137 - 1000INCOMPATIBLE WITH RFC[1000]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal SapI.rc site found at 2468
Design Notes
This BioBrick was designed to fit the RFC 10 standard using the primers:
Forward: tactagtagcggccgctgcagCCGTTATAGAAGCTTGAGTATT
Reverse: gaattcgcggccgcttctagagGCCCATGGATATATCTCCTTCT
References
1. Jacob F; Monod J. Genetic regulatory mechanisms in the synthesis of proteins, J Mol Biol. journal, 1961, vol. 3: p. 318—356.
2. J. Parker, Encyclopedia of Genetics, 2001
3. Klaus I. Matthaei, in Handbook of Stem Cells, 2004 (For picture)