Difference between revisions of "Part:BBa K3142014"
(→SZPT-China) |
(→SZPT-China) |
||
| Line 23: | Line 23: | ||
<!-- --> | <!-- --> | ||
==SZPT-China== | ==SZPT-China== | ||
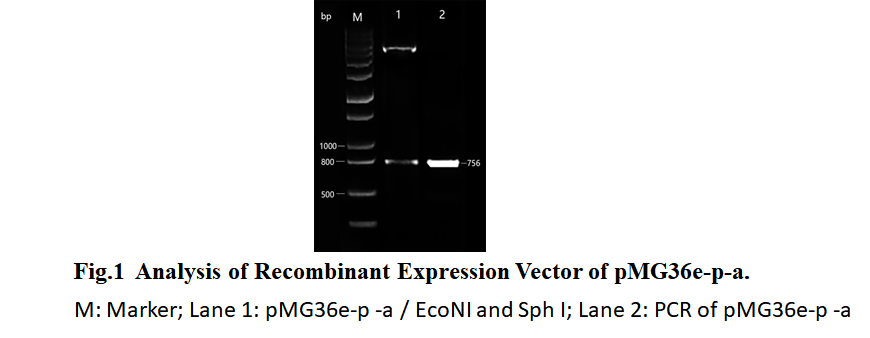
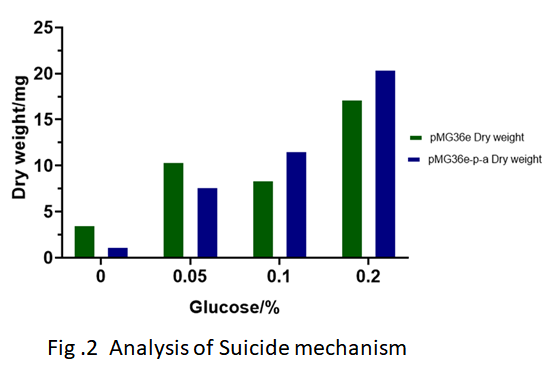
| − | To ensure that the transgenic bacteria are not harmful to the environment, we constructed the autolytic enzyme gene acmA of the lactic acid bacteria downstream of the glucose starvation promoter. The results of the construction of the recombinant plasmid pMG36e-pα-crp-acmA named pMG36e-P-a are shown in Fig. 1. The results of LAB MG1363 and LAB MG1363 with pMG36e-p - a after induction of recombinant bacteria by different glucose concentrations are shown in Fig. 2. The results showed that when the concentration of glucose in the environment was less than 0.05%, the growth of recombinant bacteria was significantly inhibited. | + | To ensure that the transgenic bacteria are not harmful to the environment, we constructed the autolytic enzyme gene acmA of the ''lactic acid bacteria'' downstream of the glucose starvation promoter. The results of the construction of the recombinant plasmid pMG36e-pα-crp-acmA named pMG36e-P-a are shown in Fig. 1. The results of ''LAB MG1363'' and ''LAB MG1363'' with pMG36e-p - a after induction of recombinant bacteria by different glucose concentrations are shown in Fig. 2. The results showed that when the concentration of glucose in the environment was less than 0.05%, the growth of recombinant bacteria was significantly inhibited. |
[[file:P-A123.png|center|500px]] | [[file:P-A123.png|center|500px]] | ||
[[file:P-A1231.png|center|500px]] | [[file:P-A1231.png|center|500px]] | ||
Revision as of 12:55, 21 October 2019
PT-αcrp + acmA
what it is:
what it does:
how to use it in their projects:
Sequence and Features
Assembly Compatibility:
- 10INCOMPATIBLE WITH RFC[10]Illegal EcoRI site found at 26
Illegal XbaI site found at 729
Illegal PstI site found at 253 - 12INCOMPATIBLE WITH RFC[12]Illegal EcoRI site found at 26
Illegal PstI site found at 253 - 21INCOMPATIBLE WITH RFC[21]Illegal EcoRI site found at 26
Illegal BamHI site found at 1
Illegal BamHI site found at 681 - 23INCOMPATIBLE WITH RFC[23]Illegal EcoRI site found at 26
Illegal XbaI site found at 729
Illegal PstI site found at 253 - 25INCOMPATIBLE WITH RFC[25]Illegal EcoRI site found at 26
Illegal XbaI site found at 729
Illegal PstI site found at 253 - 1000COMPATIBLE WITH RFC[1000]
SZPT-China
To ensure that the transgenic bacteria are not harmful to the environment, we constructed the autolytic enzyme gene acmA of the lactic acid bacteria downstream of the glucose starvation promoter. The results of the construction of the recombinant plasmid pMG36e-pα-crp-acmA named pMG36e-P-a are shown in Fig. 1. The results of LAB MG1363 and LAB MG1363 with pMG36e-p - a after induction of recombinant bacteria by different glucose concentrations are shown in Fig. 2. The results showed that when the concentration of glucose in the environment was less than 0.05%, the growth of recombinant bacteria was significantly inhibited.
Reference
- William Henry Bothfeld, Grace Kapov, and Keith Tyo. A glucose-sensing toggle switch for autonomous, high productivity genetic control. ACS Synth. 2017, 6(7):1296-1304.