Difference between revisions of "Part:BBa K3140010:Design"
| Line 7: | Line 7: | ||
===Design Notes=== | ===Design Notes=== | ||
| − | |||
| + | This part was based on the native VVD protein from (''Neurospora crassa''). The VVD36 coding sequence was generated by removing the first 36 amino acid residues and appending a start codon to the start of this truncated sequence. The cystine residue at position 73 (TGC) was then changed to alanine by changing the codon to GCC to yield VVD36-C73A. This sequence was then run through the IDT Codon Optimization Tool, with ''E. coli'' selected as the destination species. The output of this tool was used as the coding sequence for VVD36-C73A-CH4. | ||
| + | |||
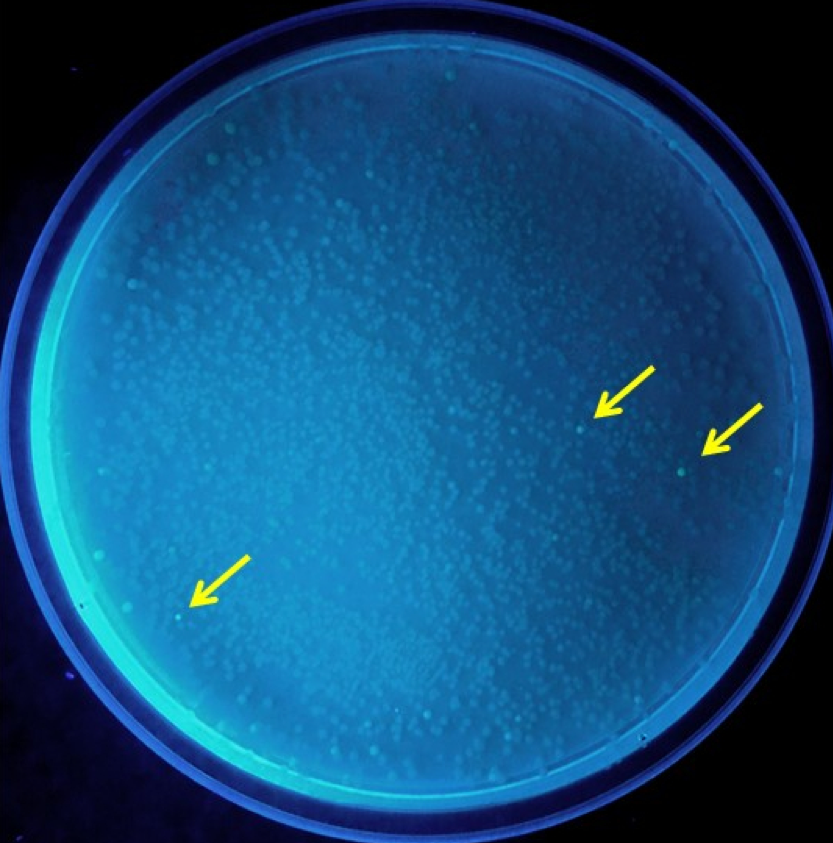
| + | Error prone PCR was performed on plasmid from the weakly fluorescent VVD36-C73A-CH4 colonies. The PCR products, when ligated in plasmids and transformed into E. coli, yielded 12 colonies that were brighter than the rest ('''Fig. 1'''). | ||
| + | |||
| + | [[Image:T--Sydney Australia--errorplate.png|frame|none|'''Fig. 1''': Results from initial ligation and transformation of error-prone PCR products into ''E. coli'' TOP10 cells. Extra bright colonies are indicated with yellow arrows.]] | ||
| + | |||
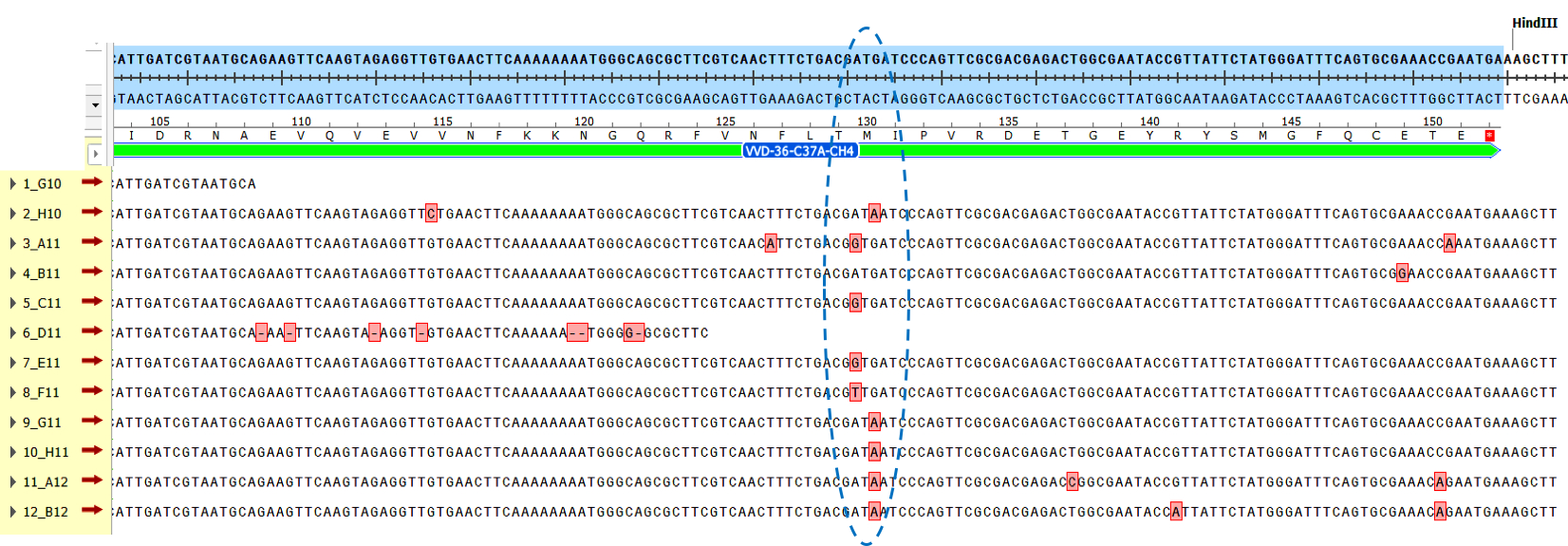
| + | The sequencing results from these bright colonies revealed that there was a common mutation in the majority of these genes ('''Fig. 2'''). This mutation modified the methionine at residue 130 to either an isoleucine (found in 5 of the mutants), a threonine (found in three of the mutants), or a leucine residue (found in one of the mutants). | ||
| + | |||
| + | [[Image:T--Sydney_Australia--errorPCR.png|900px|thumb|none|'''Fig. 2''': Sequence alignment of error-prone PCR mutants on original VVD gene, with the region of common M130 mutations circled.]] | ||
| + | |||
| + | The final coding sequence, VVD36-C73A-CH4-M130I, was designed with this methionine to isoleucine change. | ||
| + | |||
| + | This part was intended for use in pK18 as a test of our codon harmonisation model. To achieve this, the part was ordered in a gBlock flanked by EcoRI and HindIII sites to enable traditional restriction cloning into pK18. A RBS sequence was added to the gBlock upstream of the part. The RBS sequence used was the consensus Shine-Dalgarno sequence. | ||
===Source=== | ===Source=== | ||
| − | + | The native sequence for VVD (''Neurospora crassa'') was obtained from NCBI: GenBank accession [https://www.ncbi.nlm.nih.gov/nuccore/AF338412.1 AF338412.1] | |
===References=== | ===References=== | ||
| + | '''1.''' Schwerdtfeger, C. & Linden, H. VIVID is a flavoprotein and serves as a fungal blue light photoreceptor for photoadaptation. ''EMBO J'' '''22''', 4846-55 (2003). | ||
| + | '''2.''' Zoltowski, B.D. ''et al.'' Conformational switching in the fungal light sensor Vivid. ''Science'' '''316''', 1054-7 (2007). | ||
Revision as of 11:32, 21 October 2019
VVD36-C73A-CH4-M130I
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Design Notes
This part was based on the native VVD protein from (Neurospora crassa). The VVD36 coding sequence was generated by removing the first 36 amino acid residues and appending a start codon to the start of this truncated sequence. The cystine residue at position 73 (TGC) was then changed to alanine by changing the codon to GCC to yield VVD36-C73A. This sequence was then run through the IDT Codon Optimization Tool, with E. coli selected as the destination species. The output of this tool was used as the coding sequence for VVD36-C73A-CH4.
Error prone PCR was performed on plasmid from the weakly fluorescent VVD36-C73A-CH4 colonies. The PCR products, when ligated in plasmids and transformed into E. coli, yielded 12 colonies that were brighter than the rest (Fig. 1).
The sequencing results from these bright colonies revealed that there was a common mutation in the majority of these genes (Fig. 2). This mutation modified the methionine at residue 130 to either an isoleucine (found in 5 of the mutants), a threonine (found in three of the mutants), or a leucine residue (found in one of the mutants).
The final coding sequence, VVD36-C73A-CH4-M130I, was designed with this methionine to isoleucine change.
This part was intended for use in pK18 as a test of our codon harmonisation model. To achieve this, the part was ordered in a gBlock flanked by EcoRI and HindIII sites to enable traditional restriction cloning into pK18. A RBS sequence was added to the gBlock upstream of the part. The RBS sequence used was the consensus Shine-Dalgarno sequence.
Source
The native sequence for VVD (Neurospora crassa) was obtained from NCBI: GenBank accession AF338412.1
References
1. Schwerdtfeger, C. & Linden, H. VIVID is a flavoprotein and serves as a fungal blue light photoreceptor for photoadaptation. EMBO J 22, 4846-55 (2003). 2. Zoltowski, B.D. et al. Conformational switching in the fungal light sensor Vivid. Science 316, 1054-7 (2007).