Difference between revisions of "Part:BBa K3260030"
Maryeliz46 (Talk | contribs) |
|||
| Line 1: | Line 1: | ||
| + | __NOTOC__ | ||
| + | <partinfo>BBa_K3260030 short</partinfo> | ||
| + | <p>The N-terminal half of GFP with residues 1-157. Can be used to implement bimolecular fluorescence complementation (BiFC) on two proteins where dimerization is necessary for overall functionality. This nGFP is complementary to BBa_K3260028, a cGFP containing residues 158 - 238. | ||
| + | </p> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ===Usage and Biology:=== | ||
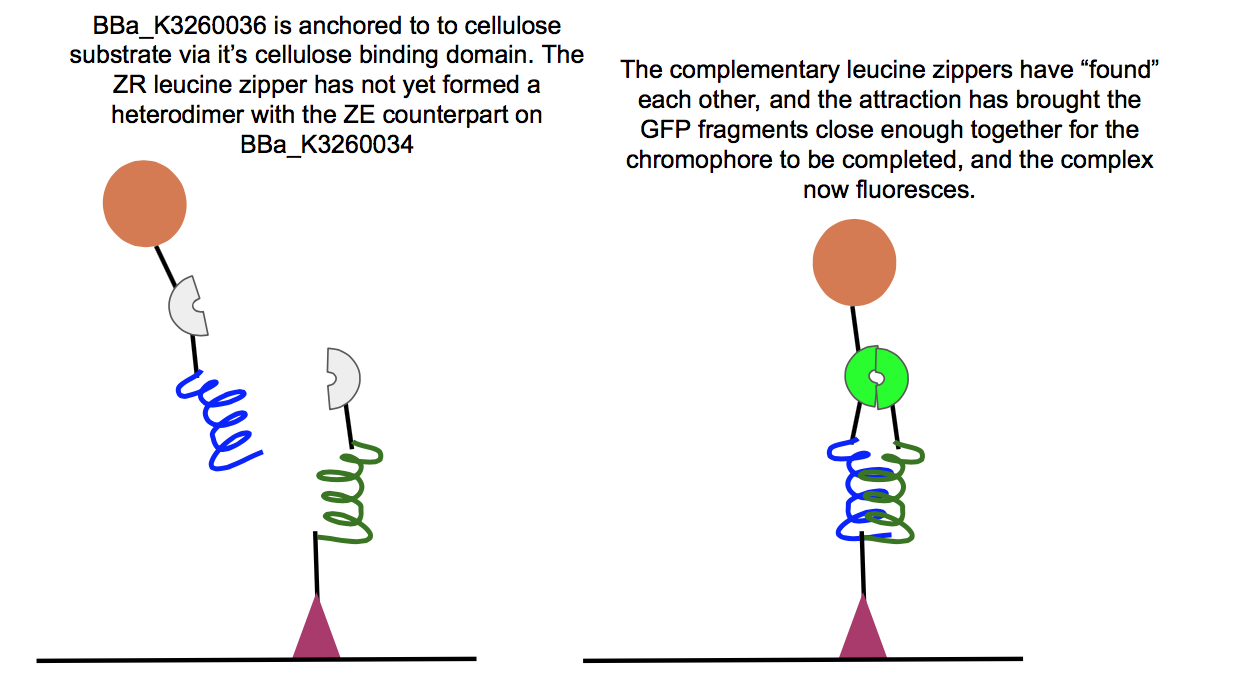
| + | <p>The position of the split was informed by Magliery et. all in their 2004 paper on GFP fragment reassembly [1]. A necessity for successful BiFC is a strong interaction between two additional domains to bring the split GFP fragments close enough to bond, completing the chromophore and fluorescing. This proximity was achieved in parts BBa_K3260036 and BBa_ K3260034, where, among other domains, each split GFP fragment was bound to heterodimeric leucine zippers, ZR leucine zipper and ZE leucine zipper respectively (BBa_K3260029 and BBa_K3260027 as basic parts). The functionality of this part is shown in a more explicit context below. | ||
| + | |||
| + | This part was used in the composite BBa_K3260034, in conjunction with the BBa_K3260036 composite. Below are the results for experiments regarding the interaction between the two composites. | ||
| + | </p> | ||
| + | |||
| + | https://2019.igem.org/wiki/images/f/f9/T--BrownStanfordPrinctn--3634_fluoresence_testing.png | ||
| + | https://2019.igem.org/wiki/images/a/a6/T--BrownStanfordPrinctn--3634_fluoresence_swatches.png | ||
| + | |||
| + | |||
| + | Above is a test of the functionality of two of our new fusion proteins, BBa_K3260036, and BBa_K3260034. If these two parts assemble correctly on the cellulose surface, there should be fluorescence, which is what we observed here. This means that the leucine zipper components of each protein aligned and held together, allowing the two halves of GFP to get close enough together to complete their chromophore and fluoresce. Because we see fluorescence (highlighted in the color swatch of each sample), the success of the leucine zippers and therefore the functionality of the last two functional domains can be inferred as well, the double cellulose binding domain anchoring the whole complex to the cellulose surface, as well as the hydrophobic domain sticking up from the N-terminal region of BBa_K3260034, and therefore the assembled protein complex. The successful assembly of these two biobricks not only shows the applicability of these constructs for paper-based microfluidics, but it also shows the potential of split-GFP reporting for fusion proteins in functional applications. | ||
| + | |||
| + | |||
| + | |||
| + | https://2019.igem.org/wiki/images/8/82/T--BrownStanfordPrinctn--3634_assembly_diagram.png | ||
| + | |||
| + | |||
| + | <p>[1] Magliery , Thomas J., and Christopher G. M. Wilson. “Detecting Protein-Protein Interactions with a Green Fluorescent Protein Fragment Reassembly Trap: Scope and Mechanism .” JACS Articles , American Chemical Society , 2 Dec. 2004.</p> | ||
| + | |||
| + | |||
| + | |||
| + | <!-- --> | ||
| + | <span class='h3bb'>Sequence and Features</span> | ||
| + | <partinfo>BBa_K3260030 SequenceAndFeatures</partinfo> | ||
| + | |||
| + | |||
| + | <!-- Uncomment this to enable Functional Parameter display | ||
| + | ===Functional Parameters=== | ||
| + | <partinfo>BBa_K3260030 parameters</partinfo> | ||
| + | <!-- --> | ||
Latest revision as of 06:54, 21 October 2019
nGFP
The N-terminal half of GFP with residues 1-157. Can be used to implement bimolecular fluorescence complementation (BiFC) on two proteins where dimerization is necessary for overall functionality. This nGFP is complementary to BBa_K3260028, a cGFP containing residues 158 - 238.
Usage and Biology:
The position of the split was informed by Magliery et. all in their 2004 paper on GFP fragment reassembly [1]. A necessity for successful BiFC is a strong interaction between two additional domains to bring the split GFP fragments close enough to bond, completing the chromophore and fluorescing. This proximity was achieved in parts BBa_K3260036 and BBa_ K3260034, where, among other domains, each split GFP fragment was bound to heterodimeric leucine zippers, ZR leucine zipper and ZE leucine zipper respectively (BBa_K3260029 and BBa_K3260027 as basic parts). The functionality of this part is shown in a more explicit context below. This part was used in the composite BBa_K3260034, in conjunction with the BBa_K3260036 composite. Below are the results for experiments regarding the interaction between the two composites.


Above is a test of the functionality of two of our new fusion proteins, BBa_K3260036, and BBa_K3260034. If these two parts assemble correctly on the cellulose surface, there should be fluorescence, which is what we observed here. This means that the leucine zipper components of each protein aligned and held together, allowing the two halves of GFP to get close enough together to complete their chromophore and fluoresce. Because we see fluorescence (highlighted in the color swatch of each sample), the success of the leucine zippers and therefore the functionality of the last two functional domains can be inferred as well, the double cellulose binding domain anchoring the whole complex to the cellulose surface, as well as the hydrophobic domain sticking up from the N-terminal region of BBa_K3260034, and therefore the assembled protein complex. The successful assembly of these two biobricks not only shows the applicability of these constructs for paper-based microfluidics, but it also shows the potential of split-GFP reporting for fusion proteins in functional applications.
[1] Magliery , Thomas J., and Christopher G. M. Wilson. “Detecting Protein-Protein Interactions with a Green Fluorescent Protein Fragment Reassembly Trap: Scope and Mechanism .” JACS Articles , American Chemical Society , 2 Dec. 2004.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
