Difference between revisions of "Part:BBa K2904022"
(→The construction of this part) |
(→Result) |
||
| Line 16: | Line 16: | ||
==<strong>Result</strong>== | ==<strong>Result</strong>== | ||
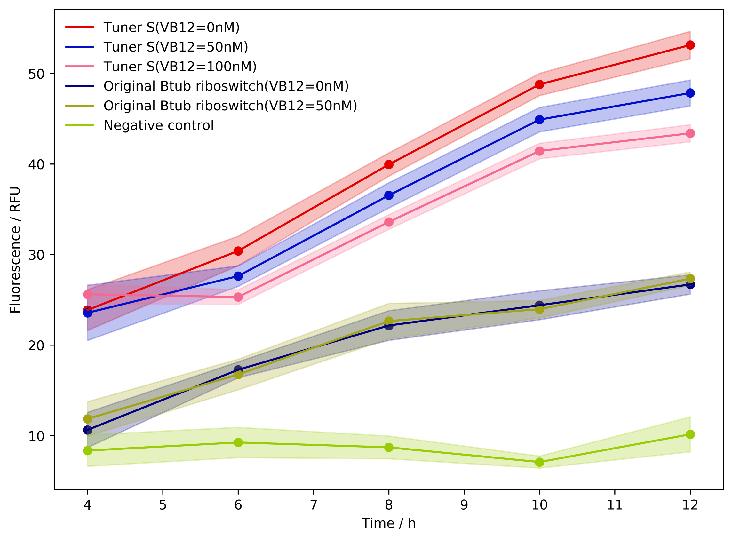
| − | We | + | We tested our system by microplate reader, which is used to reflect the intensity of sfGFP changing over time. The following chart shows the dynamic curve measured every two hours. It can prove that the modular Btub riboswitch containing SsrA degradation tag also can restore the structure of riboswitch and control the downstream gene expression during the whole cultivation period. |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
<br> | <br> | ||
</p> | </p> | ||
Revision as of 15:25, 20 October 2019
Modular Btub riboswitch containing Tuner S
Design
Background of 2019 OUC-China's project——RiboLego
Due to context-dependent performance and limited dynamic range, the widespread application of riboswitches is currently restricted. By replacing its original ORF with a new one, the structure of an aptamer domain can be subtly disrupted, resulting in a loss of ligand response. So riboswitch is still not be considered as a ‘plug and play' device. To tackle these problems, our project focuses on a standardized design principle to be used for modular and tunable riboswitch. The modular riboswitch we defined consists of the original riboswitch, Stabilizer and Tuner. Stabilizer can protect the structure of riboswitch from damage while Tuner can reduce the expression probability of fusion protein and make improvement of riboswitch function.
The construction of this part
To validate our design guideline, we employed a repressing Btub riboswitch, which can regulate the expression of TonB-dependent vitamin B12 receptor BtuB by binding adenosylcobalamin in Escherichia coli.The first 150bp of BtuB was chosen as Stabilizer of Btub riboswitch because our docking matrix suggested that a normal riboswitch structure would be observed when using this length of Stabilizer. This part was modular Btub riboswitch including the original riboswitch, Stabilizer and Tuner S. Tuner S contains SsrA degradation tag which can degrade Stabilizer, reducing the metabolic burden of cells.
Result
We tested our system by microplate reader, which is used to reflect the intensity of sfGFP changing over time. The following chart shows the dynamic curve measured every two hours. It can prove that the modular Btub riboswitch containing SsrA degradation tag also can restore the structure of riboswitch and control the downstream gene expression during the whole cultivation period.
</p>
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 156
- 1000COMPATIBLE WITH RFC[1000]