Difference between revisions of "Part:BBa K3254000"
Zongyeqing (Talk | contribs) |
Zongyeqing (Talk | contribs) |
||
| Line 3: | Line 3: | ||
<partinfo>BBa_K3254000 short</partinfo> | <partinfo>BBa_K3254000 short</partinfo> | ||
| − | This part is an improvement for the part [[Part:BBa_K2243031|BBa_K2243031]]. It can be placed between a promoter and a translational unit part and | + | This part is an improvement for the part [[Part:BBa_K2243031|BBa_K2243031]]. It can be placed between a promoter and a translational unit part and works as a normally open (NO) switch for the downstreamed gene, and switch to ON state by flipping the unidirectional terminator ECK120034435 between the att sites when it reacted with phiC31 integrase [[Part:BBa_K1039012|BBa_K1039012]]. In this improved version, two BsaI restriction site were added between attB site and terminator. As a result, it can work as a normally closed (NC) switch for the gene which was inserted between the two BsaI site and switch to OFF state when it flipped. |
| + | |||
| + | =Usage and Biology= | ||
| + | ==Visual Result as a Normally Closed Switch== | ||
| + | *We conducted a simple test to see if our design met the expection. | ||
| + | |||
| + | ===Experimental Setup=== | ||
| + | *Genetic design principle of the experimental group is described on the page of [[Part:BBa_K3254010|BBa_K3254010]]. | ||
| + | *A P15A-AmpR plasmid was co-transfered into the E.coli DH5α host cell with the reporter plasmid containing this part as the negative control. | ||
| + | *Single colonies were selected from the experimental LB-agar plate and negative control LB-agar plate, then inoculated into EP tubes with 500 μL M9 supplemented medium containing 500 μM IPTG for overnight growth at 37 °C and 200 rpm. | ||
| + | *Tubes were centrifuged at 10000g for 1 min. Then observed the GFP fluorescence of the cell precipitations under blue light. | ||
| + | |||
| + | ===Results=== | ||
| + | *IBR-C35/F55/S37/E21/T25/G22 indicate the experimental systems for phiC31/Int5/Int7/Int8/Int10/TG1 respctively. | ||
| + | *We observed the GFP fluorescence from the experimental tube as expected. | ||
| + | |||
| + | [[File:T--GENAS_China--primary_screening.png]] | ||
| + | |||
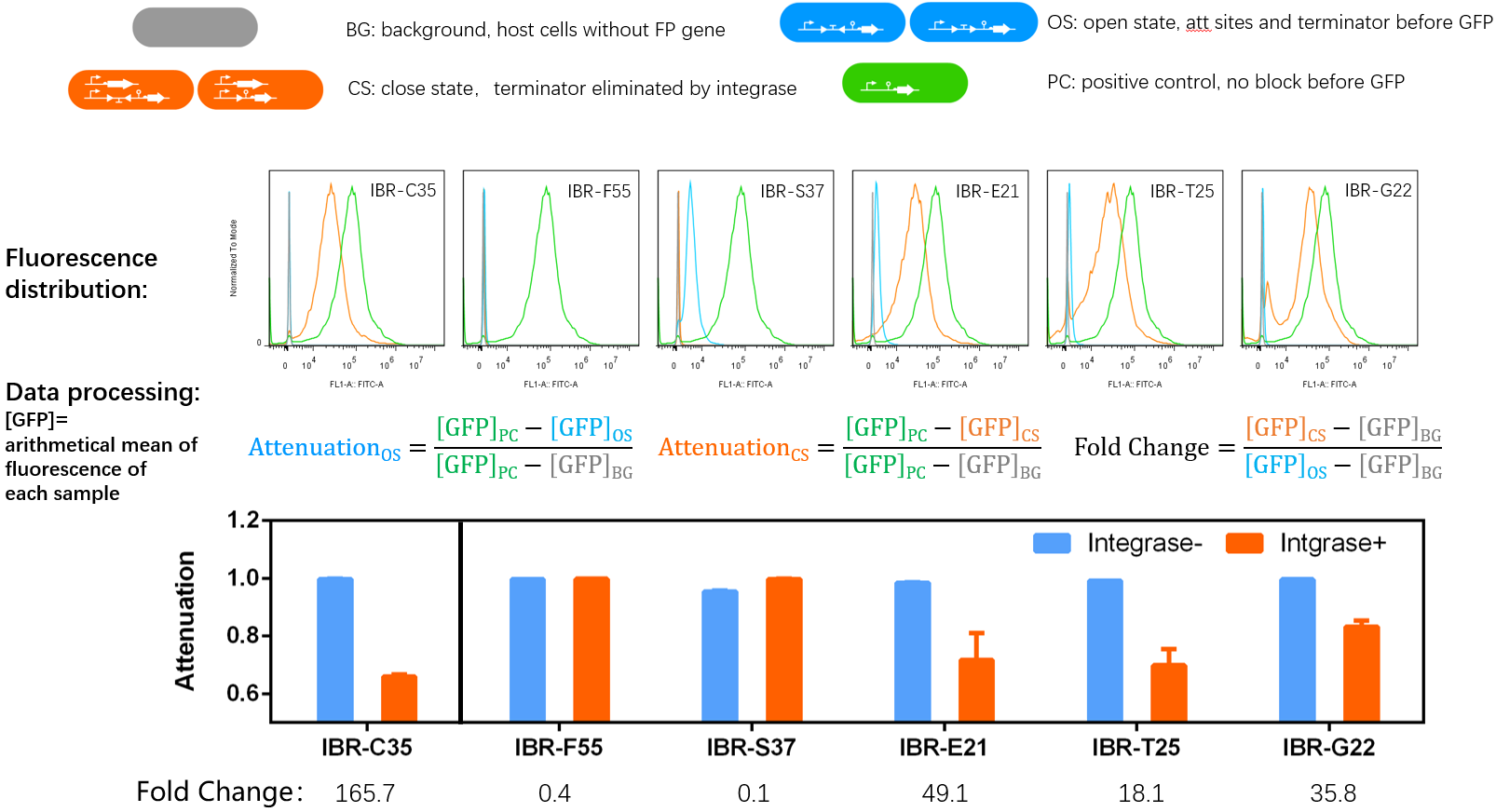
| + | ==Quantitative Characterizaion of the Normally Open Switch== | ||
| + | |||
| + | ===Experimental Setup=== | ||
| + | *Bacteria harboring the circuits (see the top part of the result image) first inoculated from single colonies into a flat-bottom 96-well plate for overnight growth. Then, the cell cultures were diluted 1000-fold with M9 supplemented medium with 500 μM IPTG inducer and growth for another 20 hours. All incubations were carried out using a Digital Thermostatic Shaker maintained at 37 °C and 1000 rpm, using flat-bottom 96-well plates sealed with sealing film. Finally, 3-μL samples each culture were transferred to a new 96-well plate containing 200 μL per well of PBS supplemented with 2 mg/mL kanamycin. | ||
| + | *The fluorescence distribution of each sample was assayed using a flow cytometry. The arithmetical mean of each sample was determined using FlowJo software. | ||
| + | *The principle of data processing is shown on the result image. | ||
| + | |||
| + | ===Results=== | ||
| + | *IBR-C35/F55/S37/E21/T25/G22 indicate the experimental systems for phiC31/Int5/Int7/Int8/Int10/TG1 respctively. | ||
| + | *Compared to other parts, this part performed well. | ||
| + | |||
| + | [[File:T--GENAS_China--primary_quantitative_characterizaion.png]] | ||
| + | |||
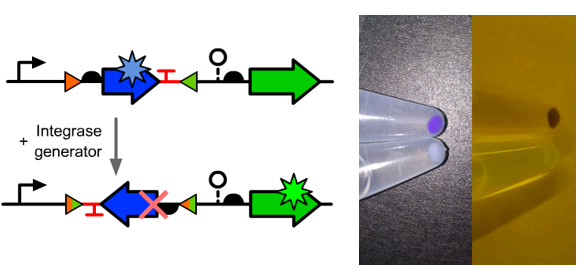
| + | ==Visual Results as a Normally Closed Switch and Toggle Switch== | ||
| + | *We inserted an [[Part:BBa_K592009|amilCP]] translational unit between the two BsaI sites. | ||
| + | *Other experiment setup were the same with "Visual Result as a Normally Closed Switch". | ||
| + | |||
| + | ===Results=== | ||
| + | *The normally open switch function well though a light blue color can be observed from the cell precipitations which might due to the incomplete diluted amilCP protein or an unexpected backward promoter. | ||
| + | *At the same time, the downstreamed GFP wasn't expression well which might due to the potential attenuation signal in the reversed amilCP sequence. | ||
| + | [[File:T--GENAS_China--NC_switch.png]] | ||
| + | |||
| + | ==Orthogonality Characterization== | ||
| + | |||
| + | ===Genetic Design=== | ||
| + | *The composition and principle of the experimental system are indicated below. | ||
| + | |||
| + | [[File:T--GENAS_China--Flip_with_backbone.PNG|200px|thumb|left| ]] | ||
| + | |||
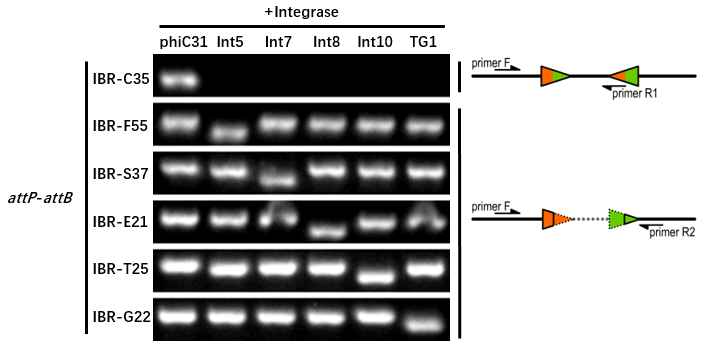
| + | ===Experimental Setup=== | ||
| + | The reporter plasmid contained this part were co-transferred into E.coli DH5α host with 6 integrase generator plasmids. | ||
| + | Then single colonies were inoculated into M9 supplemented medium for overnight growth. Then, the cell cultures were diluted 1000-fold with M9 supplemented medium with 500 μM IPTG inducer and growth for another 20 hours. All incubations were carried out using a Digital Thermostatic Shaker maintained at 37 °C and 1000 rpm, using flat-bottom 96-well plates sealed with sealing film. Finally, The cultures were sampled for genotype PCR testing. | ||
| + | The principle of genotype identification was shown on the right of results image. | ||
| + | |||
| + | ===Results=== | ||
| + | *IBR-C35 was the plasmid containing this part. | ||
| + | *The result indicates that this part can only be recombined by phiC31 integrase. | ||
| + | *The sequences after recombination are GATCAGCTCCGCGGGCAAGACCGTGCTCTTACCCAGTTGGGCGGGA (attL) and tgcgGGTGCCAGGGCGTGCCCTTGAGTTCTCTCAGTTGGGGG (attR). | ||
| + | |||
| + | [[File:T--GENAS_China--orthogonality_test.png]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| − | |||
| − | |||
| − | |||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
<partinfo>BBa_K3254000 SequenceAndFeatures</partinfo> | <partinfo>BBa_K3254000 SequenceAndFeatures</partinfo> | ||
Revision as of 11:39, 20 October 2019
phiC31attB-BsaI sites-terminator-phiC31attP(r)
This part is an improvement for the part BBa_K2243031. It can be placed between a promoter and a translational unit part and works as a normally open (NO) switch for the downstreamed gene, and switch to ON state by flipping the unidirectional terminator ECK120034435 between the att sites when it reacted with phiC31 integrase BBa_K1039012. In this improved version, two BsaI restriction site were added between attB site and terminator. As a result, it can work as a normally closed (NC) switch for the gene which was inserted between the two BsaI site and switch to OFF state when it flipped.
Usage and Biology
Visual Result as a Normally Closed Switch
- We conducted a simple test to see if our design met the expection.
Experimental Setup
- Genetic design principle of the experimental group is described on the page of BBa_K3254010.
- A P15A-AmpR plasmid was co-transfered into the E.coli DH5α host cell with the reporter plasmid containing this part as the negative control.
- Single colonies were selected from the experimental LB-agar plate and negative control LB-agar plate, then inoculated into EP tubes with 500 μL M9 supplemented medium containing 500 μM IPTG for overnight growth at 37 °C and 200 rpm.
- Tubes were centrifuged at 10000g for 1 min. Then observed the GFP fluorescence of the cell precipitations under blue light.
Results
- IBR-C35/F55/S37/E21/T25/G22 indicate the experimental systems for phiC31/Int5/Int7/Int8/Int10/TG1 respctively.
- We observed the GFP fluorescence from the experimental tube as expected.
Quantitative Characterizaion of the Normally Open Switch
Experimental Setup
- Bacteria harboring the circuits (see the top part of the result image) first inoculated from single colonies into a flat-bottom 96-well plate for overnight growth. Then, the cell cultures were diluted 1000-fold with M9 supplemented medium with 500 μM IPTG inducer and growth for another 20 hours. All incubations were carried out using a Digital Thermostatic Shaker maintained at 37 °C and 1000 rpm, using flat-bottom 96-well plates sealed with sealing film. Finally, 3-μL samples each culture were transferred to a new 96-well plate containing 200 μL per well of PBS supplemented with 2 mg/mL kanamycin.
- The fluorescence distribution of each sample was assayed using a flow cytometry. The arithmetical mean of each sample was determined using FlowJo software.
- The principle of data processing is shown on the result image.
Results
- IBR-C35/F55/S37/E21/T25/G22 indicate the experimental systems for phiC31/Int5/Int7/Int8/Int10/TG1 respctively.
- Compared to other parts, this part performed well.
Visual Results as a Normally Closed Switch and Toggle Switch
- We inserted an amilCP translational unit between the two BsaI sites.
- Other experiment setup were the same with "Visual Result as a Normally Closed Switch".
Results
- The normally open switch function well though a light blue color can be observed from the cell precipitations which might due to the incomplete diluted amilCP protein or an unexpected backward promoter.
- At the same time, the downstreamed GFP wasn't expression well which might due to the potential attenuation signal in the reversed amilCP sequence.
Orthogonality Characterization
Genetic Design
- The composition and principle of the experimental system are indicated below.
Experimental Setup
The reporter plasmid contained this part were co-transferred into E.coli DH5α host with 6 integrase generator plasmids. Then single colonies were inoculated into M9 supplemented medium for overnight growth. Then, the cell cultures were diluted 1000-fold with M9 supplemented medium with 500 μM IPTG inducer and growth for another 20 hours. All incubations were carried out using a Digital Thermostatic Shaker maintained at 37 °C and 1000 rpm, using flat-bottom 96-well plates sealed with sealing film. Finally, The cultures were sampled for genotype PCR testing. The principle of genotype identification was shown on the right of results image.
Results
- IBR-C35 was the plasmid containing this part.
- The result indicates that this part can only be recombined by phiC31 integrase.
- The sequences after recombination are GATCAGCTCCGCGGGCAAGACCGTGCTCTTACCCAGTTGGGCGGGA (attL) and tgcgGGTGCCAGGGCGTGCCCTTGAGTTCTCTCAGTTGGGGG (attR).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 59
Illegal BsaI.rc site found at 47