Difference between revisions of "Part:BBa K2997012"
(→Characterization) |
|||
| Line 5: | Line 5: | ||
===Characterization=== | ===Characterization=== | ||
| − | This year, we characterized FNR promoter ([[Part: BBa_K1123000|BBa_K1123000]]) which is an anaerobic promoter. Originally, we used this promoter to drive the expression of TAL constructs. However, finding that this promoter has a higher expression level of GFP under aerobic conditions, we did not use this promoter for our project. Instead, we used the strong constitutive promoter J23100 as our promoter. | + | This year, we characterized FNR promoter ([[Part:BBa_K1123000|BBa_K1123000]]) which is an anaerobic promoter. Originally, we used this promoter to drive the expression of TAL constructs. However, finding that this promoter has a higher expression level of GFP under aerobic conditions, we did not use this promoter for our project. Instead, we used the strong constitutive promoter J23100 as our promoter. |
We first cultured DH5a pSB1C3-P<sub>FNR</sub>-GFP under aerobic and anaerobic conditions. After 10 hours of incubation, we fixed the <i>E. coli</i> cells in 2% agarose gel and placed it on a glass slide, then place the glass slide under a microscope for image capturing. | We first cultured DH5a pSB1C3-P<sub>FNR</sub>-GFP under aerobic and anaerobic conditions. After 10 hours of incubation, we fixed the <i>E. coli</i> cells in 2% agarose gel and placed it on a glass slide, then place the glass slide under a microscope for image capturing. | ||
Revision as of 09:08, 20 October 2019
Pfnr-GFP
Characterization
This year, we characterized FNR promoter (BBa_K1123000) which is an anaerobic promoter. Originally, we used this promoter to drive the expression of TAL constructs. However, finding that this promoter has a higher expression level of GFP under aerobic conditions, we did not use this promoter for our project. Instead, we used the strong constitutive promoter J23100 as our promoter.
We first cultured DH5a pSB1C3-PFNR-GFP under aerobic and anaerobic conditions. After 10 hours of incubation, we fixed the E. coli cells in 2% agarose gel and placed it on a glass slide, then place the glass slide under a microscope for image capturing.
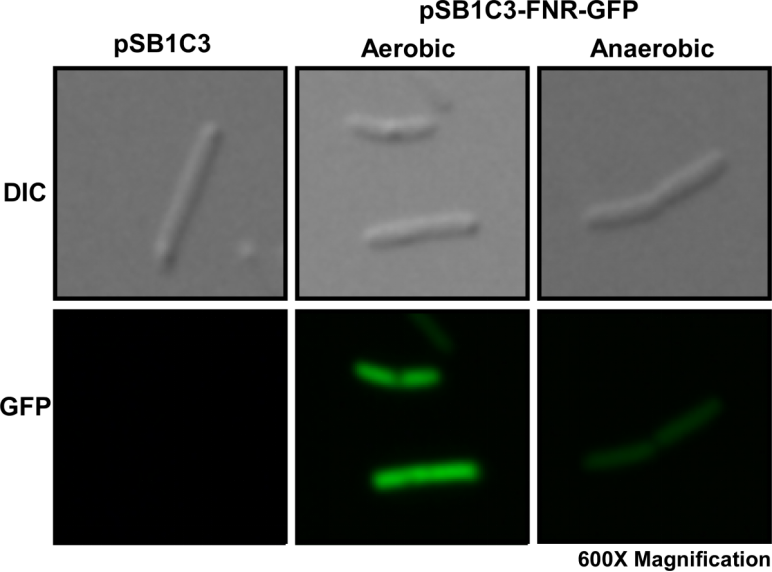
Fig. 1 Differential interference contrast (DIC) microscope image and fluorescence microscope image for a single E. coli cell.
We then compared the brightness of a single E. coli cell under a fluorescence microscope using ImageJ Intensity Processing. The results are shown below.

Fig. 2. Quantification of single E. coli cell (n=3) fluorescence intensity using ImageJ Intensity Processing after 10 hours of incubation in both aerobic and anaerobic conditions.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 39
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 790
