Difference between revisions of "Part:BBa K2997011"
| Line 4: | Line 4: | ||
===Background=== | ===Background=== | ||
| − | Tyrosine ammonia-lyase (TAL) is an enzyme that converts tyrosine into p-Coumaric acid. We engineered E. coli Nissle to express TAL to turn the excess tyrosine inside the gut into p-Coumaric acid, and compare the different constructs for improvement, BBa_K2997009 and BBa_K2997010. | + | Tyrosine ammonia-lyase (TAL) is an enzyme that converts tyrosine into <i>p</i>-Coumaric acid. We engineered <i>E. coli</i> Nissle to express TAL to turn the excess tyrosine inside the gut into <i>p</i>-Coumaric acid, and compare the different constructs for improvement, [[Part:BBa_K2997009 | BBa_K2997009]] and BBa_K2997010. |
===Expression in <i>E.coli</i>=== | ===Expression in <i>E.coli</i>=== | ||
| − | We constructed Tyrosine Ammonia Lyase with B0034 RBS , and transformed the plasmid into E. coli Nissle 1917 and confirmed it by double digestion. The results are as follows: | + | We constructed Tyrosine Ammonia Lyase with B0034 RBS , and transformed the plasmid into <i>E. coli</i> Nissle 1917 and confirmed it by double digestion. The results are as follows: |
<html> | <html> | ||
<br> | <br> | ||
| Line 19: | Line 19: | ||
Then, we carried out SDS-PAGE to check the protein expression of TAL, and the expected protein size of TAL is 54kDa. As seen in the results below, however, there’s no distinguishable band around both sizes. | Then, we carried out SDS-PAGE to check the protein expression of TAL, and the expected protein size of TAL is 54kDa. As seen in the results below, however, there’s no distinguishable band around both sizes. | ||
| + | |||
<html> | <html> | ||
<br> | <br> | ||
| Line 26: | Line 27: | ||
<br> | <br> | ||
</html> | </html> | ||
| − | Fig. 2. 12% SDS PAGE of E. coli Nissle 1917 with different plasmids. M: Marker; Lane 1: Wild Type; Lane 2: pSB1C3; Lane 3: BBa_K2997009 ; Lane 4: BBa_K2997010; Lane 5: Dual plasmid containing BBa_K2997009 and BBa_K2997000; Lane 6: Dual plasmid containing BBa_K2997010 and BBa_K2997000; Lane 7: Positive control (c.d. 3392) | + | Fig. 2. 12% SDS PAGE of <i>E. coli</i> Nissle 1917 with different plasmids. M: Marker; Lane 1: Wild Type; Lane 2: pSB1C3; Lane 3: [[Part:BBa_K2997009 | BBa_K2997009]] ; Lane 4: BBa_K2997010; Lane 5: Dual plasmid containing [[Part:BBa_K2997009 | BBa_K2997009]] and [[Part:BBa_K2997000 | BBa_K2997000]]; Lane 6: Dual plasmid containing [[Part:BBa_K2997010 | BBa_K2997010]] and [[Part:BBa_K2997000 | BBa_K2997000]]; Lane 7: Positive control (c.d. 3392) |
===RT-PCR=== | ===RT-PCR=== | ||
| − | RT-PCR experiment was used to confirm that the constructed TAL Biobrick is being transcribed. As seen in Fig.9, cDNA for both bacteria carrying TAL constructs are being detected by PCR. Confirming that the TAL genes is actually being transcribed in E. coli Nissle. | + | RT-PCR experiment was used to confirm that the constructed TAL Biobrick is being transcribed. As seen in Fig.9, cDNA for both bacteria carrying TAL constructs are being detected by PCR. Confirming that the TAL genes is actually being transcribed in <i>E. coli</i> Nissle. |
<html> | <html> | ||
<br> | <br> | ||
| Line 48: | Line 49: | ||
<br> | <br> | ||
</html> | </html> | ||
| − | Table 1. Description of templates and primers being used in each lane. | + | Table 1. Description of templates and primers being used in each lane. |
===TyrP and TAL Assay=== | ===TyrP and TAL Assay=== | ||
| − | To confirm the protein activity of TAL and TyrP, we performed a functional test using n-octanol extraction method | + | |
| + | To confirm the protein activity of TAL and TyrP, we performed a functional test using <i>n</i>-octanol extraction method,[https://2019.igem.org/Team:NCKU_Tainan/Protocols TyrP and TAL Assay], which was previously proposed by iGEM Uppsala 2013 and has been verified by HPLC<sup>[1]</sup>. The <i>p</i>-Coumaric acid concentration was measured through the absorbance value at 310nm wavelength under Nanodrop UV-Vis wavelength. The standard curve of <i>p</i>-Coumaric acid was drawn in Fig.4 to determine the relationship between <i>p</i>-Coumaric acid concentrations and its 310nm arbitrary unit (a.u). Our samples with TAL constructs were then mapped onto the standard curve, to know how much <i>p</i>-Coumaric acid is being produced. | ||
| + | |||
<html> | <html> | ||
<br> | <br> | ||
| Line 59: | Line 62: | ||
<br> | <br> | ||
</html> | </html> | ||
| − | Fig. 4. The standard curve of p-Coumaric acid concentration in correlation with absorbance at 310nm, which is provided by genetic E. coli Nissle in LB broth after 48 hours. | + | Fig. 4. The standard curve of <i>p</i>-Coumaric acid concentration in correlation with absorbance at 310nm, which is provided by genetic <i>E. coli</i> Nissle in LB broth after 48 hours. |
| − | We compared the TAL constructs containing the native and B0034 ribosome binding sites (BBa_K2997010), to determine if p-Coumaric Acid production is improved by changing the ribosome binding sites. From the results seen in Fig. 5, BBa_K2997010 is able to produce a higher amount of p-Coumaric acid. Hence, we are able to prove that by changing the RBS (from Native to B0034), the conversion of tyrosine into p-Coumaric acid can increase by 1.73-fold. | + | We compared the TAL constructs containing the native and B0034 ribosome binding sites (BBa_K2997010), to determine if <i>p</i>-Coumaric Acid production is improved by changing the ribosome binding sites. From the results seen in Fig. 5, BBa_K2997010 is able to produce a higher amount of <i>p</i>-Coumaric acid. Hence, we are able to prove that by changing the RBS (from Native to B0034), the conversion of tyrosine into <i>p</i>-Coumaric acid can increase by 1.73-fold. |
<html> | <html> | ||
<br> | <br> | ||
| Line 69: | Line 72: | ||
<br> | <br> | ||
</html> | </html> | ||
| − | Fig. 5. p-Coumaric acid/O.D.600 levels of E. coli Nissle with TAL and tyrP in LB with 1mM tyrosine | + | Fig. 5. <i>p</i>-Coumaric acid/O.D.600 levels of <i>E. coli</i> Nissle with TAL and <i>tyrP</i> in LB with 1mM tyrosine |
| − | To further prove that the TAL enzyme can specifically use tyrosine as its substrate, we cultured E. coli Nissle with dual plasmids containing TAL with B0034 RBS (BBa_K2997010) and tyrosine transporter (BBa_K2997009) in LB medium with different concentrations of tyrosine with hopes to see a dose-dependent effect. As seen in Fig. 6, although no significance in p-Coumaric acid production in culture supplemented with 0.5mM and 1.0mM tyrosine was detected, there was an increasing trend. Furthermore, there was a significant increase when comparing culture supplemented with 1.0mM and 2.0mM tyrosine. We speculate that there was indeed a dose-dependent effect, but due to the sensitivity limitations of n-octanol extraction method, it was not apparent. | + | To further prove that the TAL enzyme can specifically use tyrosine as its substrate, we cultured <i>E. coli</i> Nissle with dual plasmids containing TAL with B0034 RBS (BBa_K2997010) and tyrosine transporter ([[Part:BBa_K2997009 | BBa_K2997009]]) in LB medium with different concentrations of tyrosine with hopes to see a dose-dependent effect. As seen in Fig. 6, although no significance in <i>p</i>-Coumaric acid production in culture supplemented with 0.5mM and 1.0mM tyrosine was detected, there was an increasing trend. Furthermore, there was a significant increase when comparing culture supplemented with 1.0mM and 2.0mM tyrosine. We speculate that there was indeed a dose-dependent effect, but due to the sensitivity limitations of <i>n</i>-octanol extraction method, it was not apparent. |
<html> | <html> | ||
<br> | <br> | ||
| Line 79: | Line 82: | ||
<br> | <br> | ||
</html> | </html> | ||
| − | Fig 6. p-Coumaric acid/O.D. 600 production levels from E. coli Nissle with dual plasmids of tyrP and TAL with B0034 RBS in LB with different concentrations of tyrosine. | + | Fig 6. <i>p</i>-Coumaric acid/O.D. 600 production levels from <i>E. coli</i> Nissle with dual plasmids of <i>tyrP</i> and TAL with B0034 RBS in LB with different concentrations of tyrosine. |
| Line 94: | Line 97: | ||
<partinfo>BBa_K2997011 parameters</partinfo> | <partinfo>BBa_K2997011 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| + | |||
| + | ===Reference=== | ||
| + | [1] http://2013.igem.org/Team:Uppsala | ||
Revision as of 08:58, 20 October 2019
B0034-sam8
Background
Tyrosine ammonia-lyase (TAL) is an enzyme that converts tyrosine into p-Coumaric acid. We engineered E. coli Nissle to express TAL to turn the excess tyrosine inside the gut into p-Coumaric acid, and compare the different constructs for improvement, BBa_K2997009 and BBa_K2997010.
Expression in E.coli
We constructed Tyrosine Ammonia Lyase with B0034 RBS , and transformed the plasmid into E. coli Nissle 1917 and confirmed it by double digestion. The results are as follows:
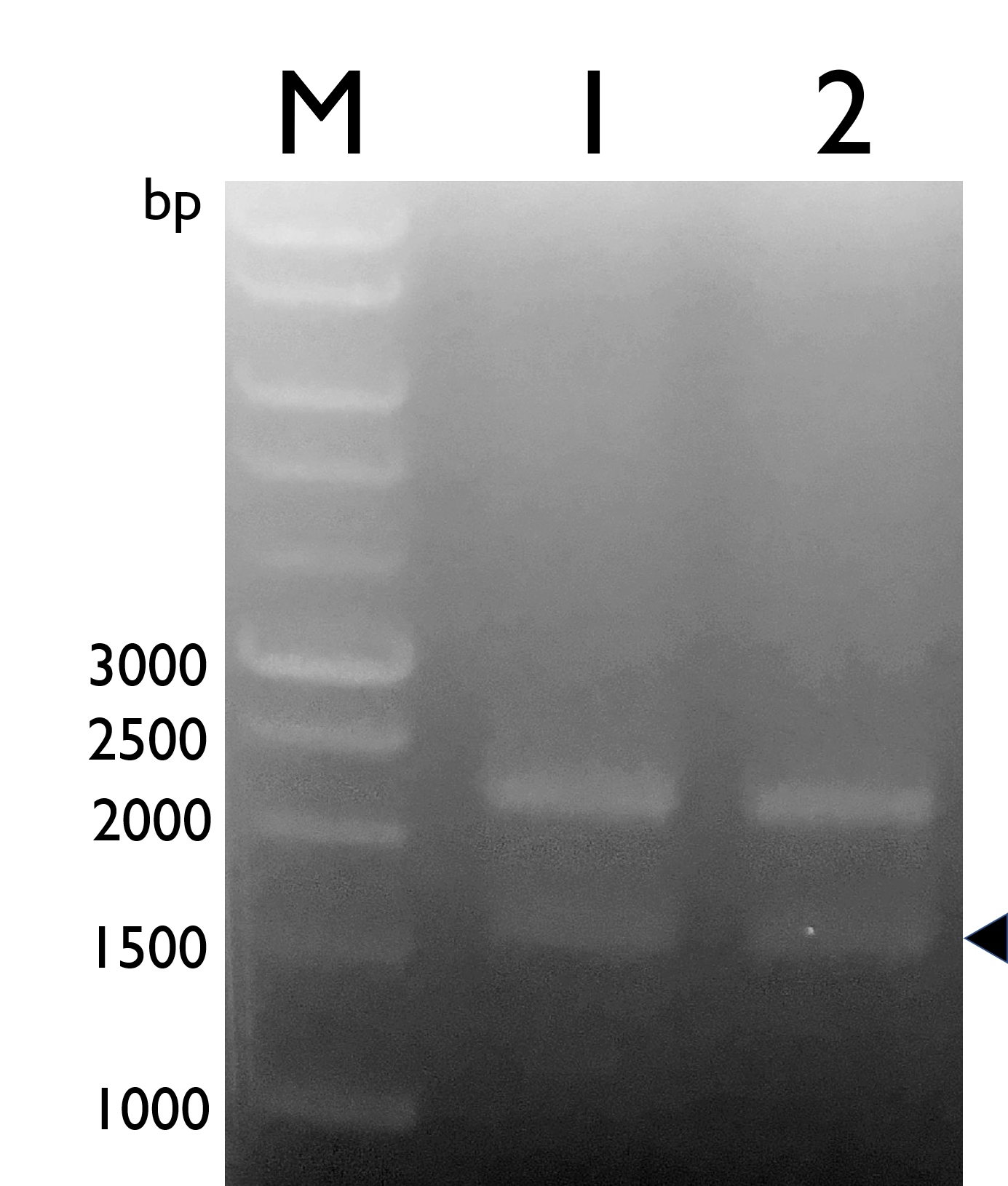
Fig. 1.Confirmation of BBa_K2997010 by double digestion, arrow indicates TAL with B0034 (~1600bp).
Then, we carried out SDS-PAGE to check the protein expression of TAL, and the expected protein size of TAL is 54kDa. As seen in the results below, however, there’s no distinguishable band around both sizes.
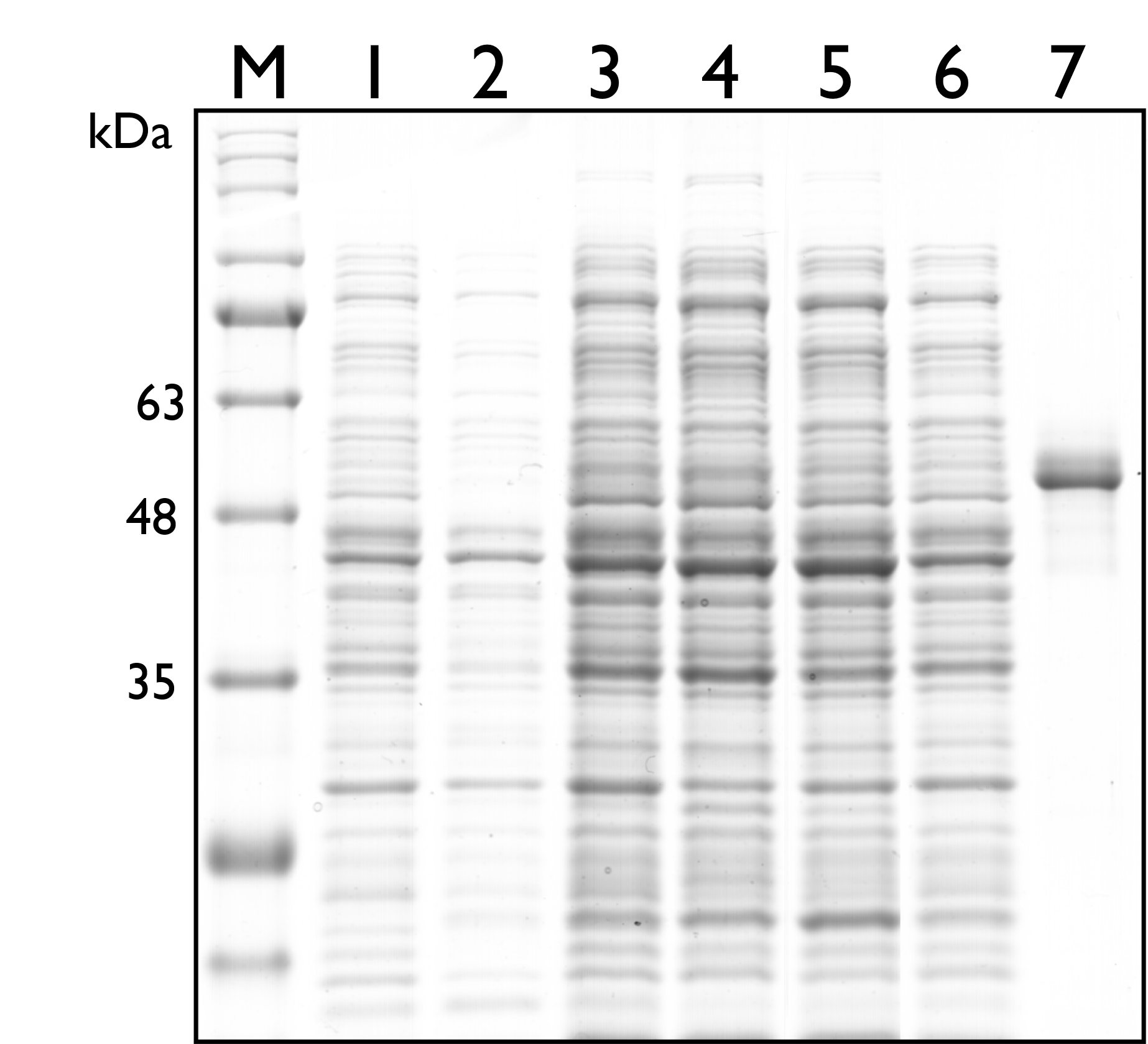
Fig. 2. 12% SDS PAGE of E. coli Nissle 1917 with different plasmids. M: Marker; Lane 1: Wild Type; Lane 2: pSB1C3; Lane 3: BBa_K2997009 ; Lane 4: BBa_K2997010; Lane 5: Dual plasmid containing BBa_K2997009 and BBa_K2997000; Lane 6: Dual plasmid containing BBa_K2997010 and BBa_K2997000; Lane 7: Positive control (c.d. 3392)
RT-PCR
RT-PCR experiment was used to confirm that the constructed TAL Biobrick is being transcribed. As seen in Fig.9, cDNA for both bacteria carrying TAL constructs are being detected by PCR. Confirming that the TAL genes is actually being transcribed in E. coli Nissle.

Fig. 3. Reverse Transcription(RT)-PCR Results to confirm that our construct is being transcribed. (a) Schematics show location of amplified regions and primers. (b) 1.5% Agarose gel shows PCR results. All products have expected size of 250bp as shown in (a). (cDNA: Total cDNA; RNA: Total RNA; plasmid: pSB1C3 containing respective Biobrick.)

Table 1. Description of templates and primers being used in each lane.
TyrP and TAL Assay
To confirm the protein activity of TAL and TyrP, we performed a functional test using n-octanol extraction method,TyrP and TAL Assay, which was previously proposed by iGEM Uppsala 2013 and has been verified by HPLC[1]. The p-Coumaric acid concentration was measured through the absorbance value at 310nm wavelength under Nanodrop UV-Vis wavelength. The standard curve of p-Coumaric acid was drawn in Fig.4 to determine the relationship between p-Coumaric acid concentrations and its 310nm arbitrary unit (a.u). Our samples with TAL constructs were then mapped onto the standard curve, to know how much p-Coumaric acid is being produced.
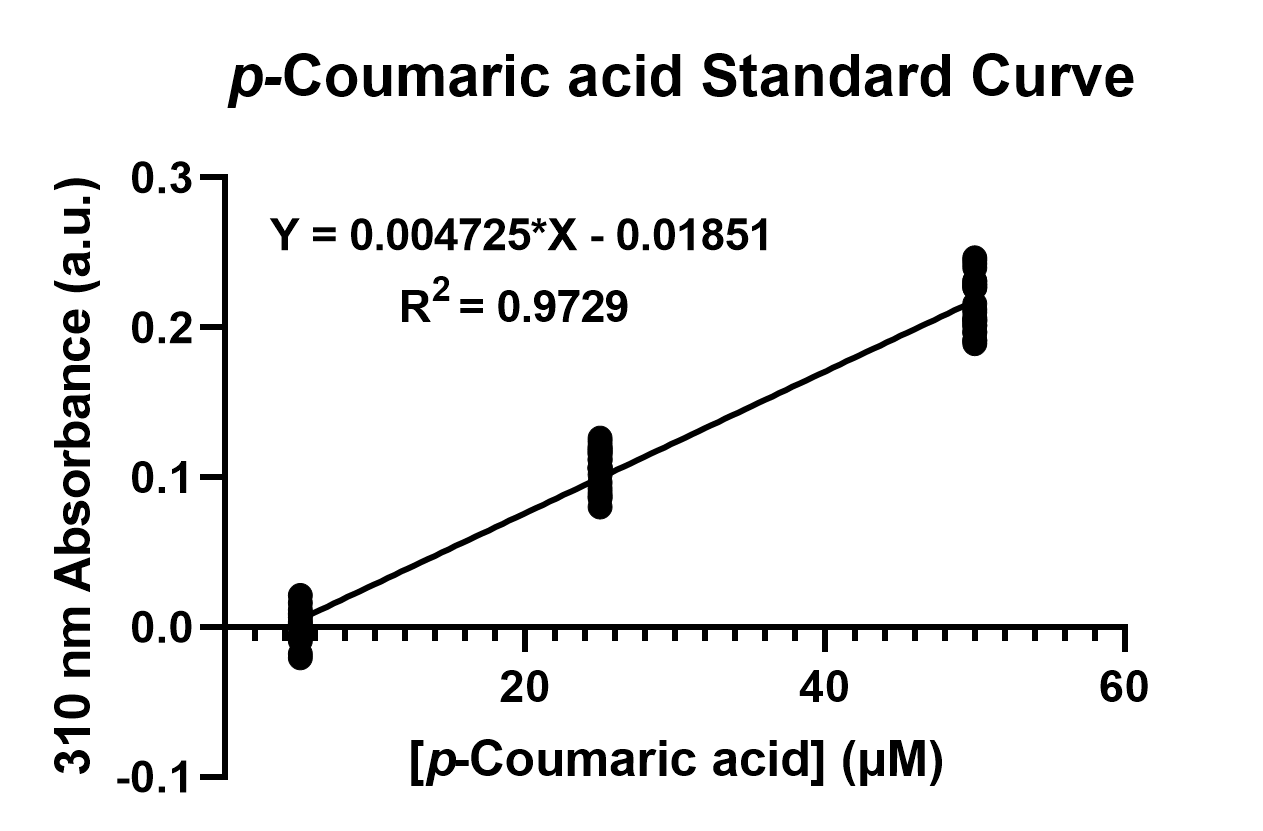
Fig. 4. The standard curve of p-Coumaric acid concentration in correlation with absorbance at 310nm, which is provided by genetic E. coli Nissle in LB broth after 48 hours.
We compared the TAL constructs containing the native and B0034 ribosome binding sites (BBa_K2997010), to determine if p-Coumaric Acid production is improved by changing the ribosome binding sites. From the results seen in Fig. 5, BBa_K2997010 is able to produce a higher amount of p-Coumaric acid. Hence, we are able to prove that by changing the RBS (from Native to B0034), the conversion of tyrosine into p-Coumaric acid can increase by 1.73-fold.
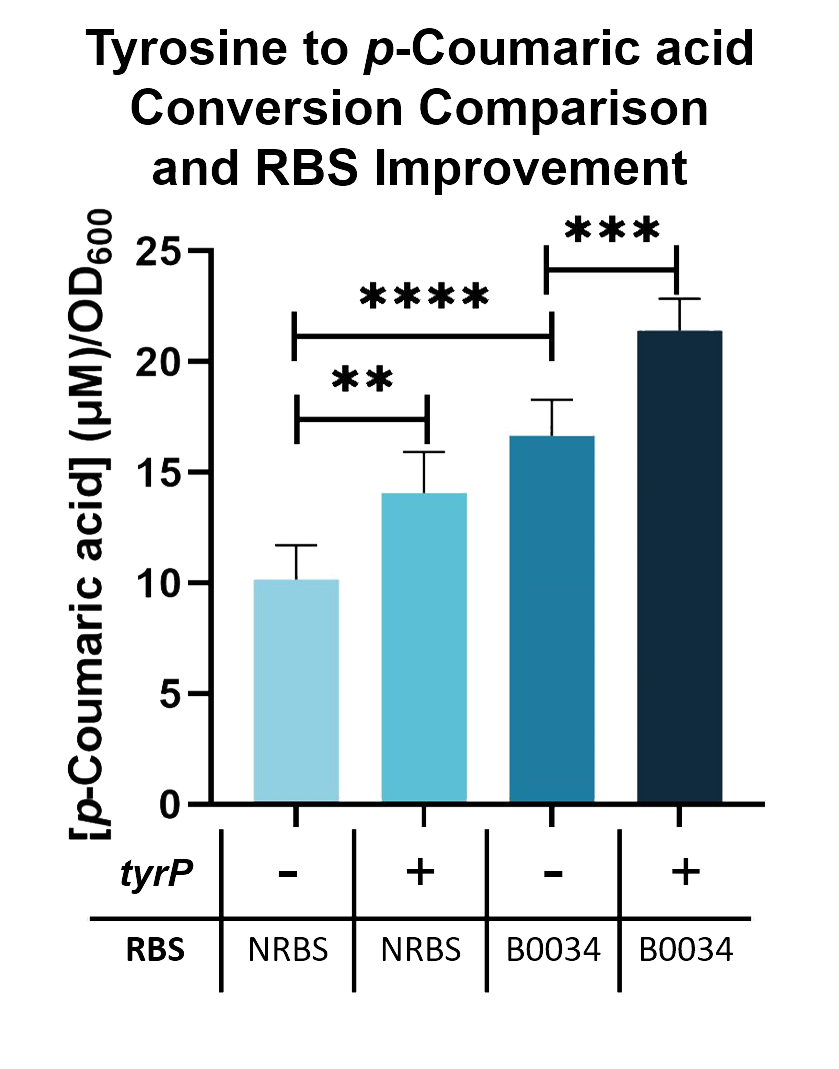
Fig. 5. p-Coumaric acid/O.D.600 levels of E. coli Nissle with TAL and tyrP in LB with 1mM tyrosine
To further prove that the TAL enzyme can specifically use tyrosine as its substrate, we cultured E. coli Nissle with dual plasmids containing TAL with B0034 RBS (BBa_K2997010) and tyrosine transporter ( BBa_K2997009) in LB medium with different concentrations of tyrosine with hopes to see a dose-dependent effect. As seen in Fig. 6, although no significance in p-Coumaric acid production in culture supplemented with 0.5mM and 1.0mM tyrosine was detected, there was an increasing trend. Furthermore, there was a significant increase when comparing culture supplemented with 1.0mM and 2.0mM tyrosine. We speculate that there was indeed a dose-dependent effect, but due to the sensitivity limitations of n-octanol extraction method, it was not apparent.

Fig 6. p-Coumaric acid/O.D. 600 production levels from E. coli Nissle with dual plasmids of tyrP and TAL with B0034 RBS in LB with different concentrations of tyrosine.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 828
Illegal BamHI site found at 1230 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 571
Illegal AgeI site found at 518
Illegal AgeI site found at 687 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 122
Illegal BsaI site found at 388
Reference
[1] http://2013.igem.org/Team:Uppsala
