Difference between revisions of "Part:BBa K3257069"
| Line 4: | Line 4: | ||
This degradation tag has the amino acid sequence of AANDENYALAA. When fused after a protein, it will cause protein degradation to happen. | This degradation tag has the amino acid sequence of AANDENYALAA. When fused after a protein, it will cause protein degradation to happen. | ||
Using EGFP (BBa_E0040 https://parts.igem.org/Part:BBa_E0040) as a reporter protein, we have measured the fluorescent intensity of EGFP attached to different degradation tags at the steady-state (Figure 1), which indicates the degradation rate of each degradation tag when attached after a CDS of a protein. | Using EGFP (BBa_E0040 https://parts.igem.org/Part:BBa_E0040) as a reporter protein, we have measured the fluorescent intensity of EGFP attached to different degradation tags at the steady-state (Figure 1), which indicates the degradation rate of each degradation tag when attached after a CDS of a protein. | ||
| + | |||
[[File:Degradation tags-steady state.png|center|500px|thumb|'''Figure 1. Degradation tag greatly reduces the protein level at steady state. WT represents the positive control of EGFP without any tag attachment. The five degradation tags are represented by their last five amino acid sequence. The vertical axis shows the quantitative analysis of EGFP fluorescence (excitation wavelength: 485 nm; detection wavelength: 528 nm), normalized by cell amount (OD600). The fluorescence is quantified by the concentration of green fluorescein, cell number is quantified by the number of silicon beads, both are from the distributed measurement kit. Fluorescence below detection level is eliminated. Error bar stands for the SEM of 3 replicates. t-test is performed between WT and each degradation tag, P<0.0001 (****).]] | [[File:Degradation tags-steady state.png|center|500px|thumb|'''Figure 1. Degradation tag greatly reduces the protein level at steady state. WT represents the positive control of EGFP without any tag attachment. The five degradation tags are represented by their last five amino acid sequence. The vertical axis shows the quantitative analysis of EGFP fluorescence (excitation wavelength: 485 nm; detection wavelength: 528 nm), normalized by cell amount (OD600). The fluorescence is quantified by the concentration of green fluorescein, cell number is quantified by the number of silicon beads, both are from the distributed measurement kit. Fluorescence below detection level is eliminated. Error bar stands for the SEM of 3 replicates. t-test is performed between WT and each degradation tag, P<0.0001 (****).]] | ||
Revision as of 18:53, 19 October 2019
SsrA(LAA)
This degradation tag has the amino acid sequence of AANDENYALAA. When fused after a protein, it will cause protein degradation to happen. Using EGFP (BBa_E0040 https://parts.igem.org/Part:BBa_E0040) as a reporter protein, we have measured the fluorescent intensity of EGFP attached to different degradation tags at the steady-state (Figure 1), which indicates the degradation rate of each degradation tag when attached after a CDS of a protein.
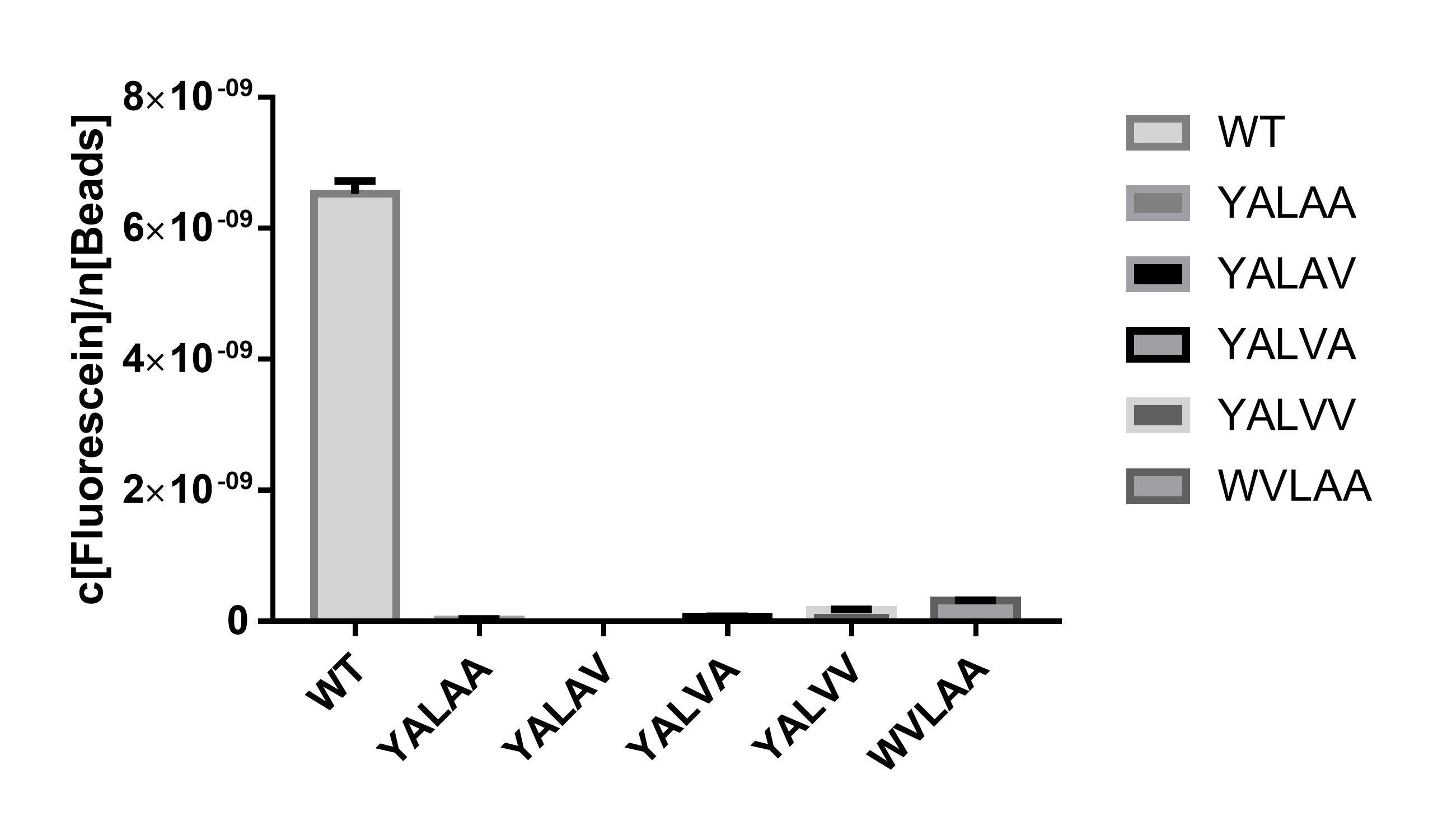
Figure 1. Degradation tag greatly reduces the protein level at steady state. WT represents the positive control of EGFP without any tag attachment. The five degradation tags are represented by their last five amino acid sequence. The vertical axis shows the quantitative analysis of EGFP fluorescence (excitation wavelength: 485 nm; detection wavelength: 528 nm), normalized by cell amount (OD600). The fluorescence is quantified by the concentration of green fluorescein, cell number is quantified by the number of silicon beads, both are from the distributed measurement kit. Fluorescence below detection level is eliminated. Error bar stands for the SEM of 3 replicates. t-test is performed between WT and each degradation tag, P<0.0001 (****).
Sequence and Features
Assembly Compatibility:
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
