Difference between revisions of "Part:BBa K1378001"
Cornelligem (Talk | contribs) m |
|||
| Line 17: | Line 17: | ||
</html> | </html> | ||
| − | |||
===Usage and Biology=== | ===Usage and Biology=== | ||
| − | |||
| − | |||
| − | |||
=='''Characterization'''== | =='''Characterization'''== | ||
| Line 52: | Line 48: | ||
</html> | </html> | ||
| + | |||
| + | ==shanghai-HS 2019's characterization== | ||
| + | ===BBa_K1378001 MlrA=== | ||
| + | =='''Methods:'''== | ||
| + | <p>1. replicating the MlrA DNA through a technology called PCR, Gel Electrophoresis and Extraction of MlrA</p> | ||
| + | <p>2. combining the gene MlrA with plasmids, and transformed the plasmids to DH5-alpha, a strain of E. coli.</p> | ||
| + | <p>3. Senting the extracted plasmid to Sangon, a biotech company, for DNA sequencing after a series of procedures.</p> | ||
| + | <p>4. transforming the plasmid to E. coli BL21(DE3), another type of E-coli characterized in producing protein, to express the targeted protein.</p> | ||
| + | <p>5. purifing the protein MlrA and utilizing HPLC to determine the effectiveness of the protein MlrA.</p> | ||
| + | |||
| + | =='''Results:'''== | ||
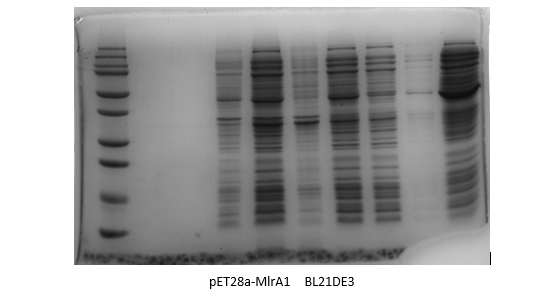
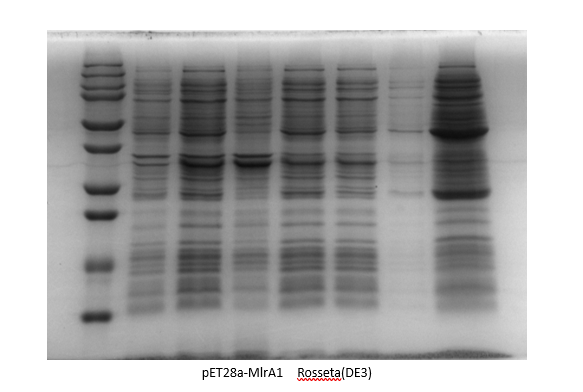
| + | <p>By adding the protein that binds to the Ni21-NTA to the affinity column, we were able to collect the liquid that went through. We ran the SDS-Page Gel with thirteen chosen samples to verify the results of the protein purification. It turns out, we successfully purified pET28-mlrA1, indiciating that it can be devoted to degrading MC-LR.</p> | ||
| + | [[Image:bl21de3.png|400px|center|Characterization of popular BioBrick RBSs]] | ||
| + | |||
| + | [[Image:rosseta(de3).png|400px|center|Characterization of popular BioBrick RBSs]] | ||
| + | |||
| + | =='''Results:'''== | ||
| + | ===Protein Purification=== | ||
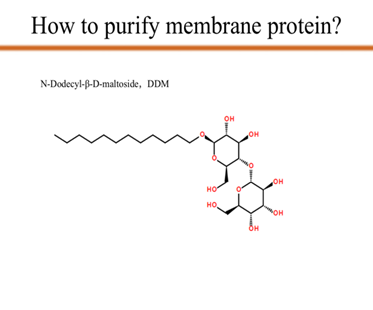
| + | <p>As you can see from the picture, we use N-Dodecyl-β-D-maltoside, which is also known as DDM, to transfer the target membrane protein mlrA from the supernatant to the membrane of DDM. Since DDM provides the membrane which mlrA depends on, with the DDM concentration increases, the concentration of the mlrA in the supernatant decreases to zero. That is the last step of protein purification.</p> | ||
| + | [[Image:k1378001-1.png|400px|center|Characterization of popular BioBrick RBSs]] | ||
| + | <p>After centrifuge the solution, to further purify the protein, we put Ni21-NTA with the supernatant and incubate overnight. The mlrA will be combined with the specific binding sites on Ni21-NTA. Then, using the method of gradient elution, while the amount of imidazole, which is a competitive agent in this step, increases, it will compete with mlrA on the binding sites of the Ni column. Therefore, the mlrA is gradually elated.</p> | ||
| + | |||
| + | ===Enzyme Function Test=== | ||
| + | ====High Performance Liquid Chromatography (HPLC)==== | ||
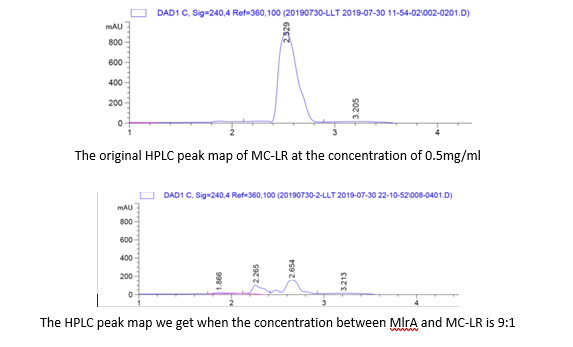
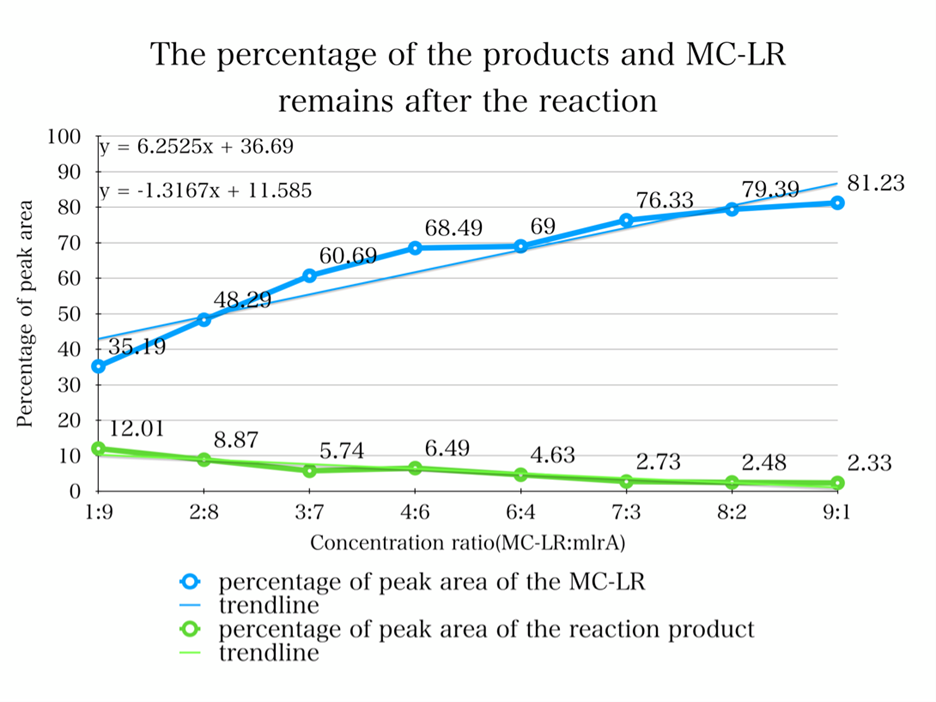
| + | <p>To quantify whether MC-LR can be degraded from the samples, we conducted a control variable experiments, and the following are the data we got:</p> | ||
| + | [[Image:k1378001-2.png|400px|center|Characterization of popular BioBrick RBSs]] | ||
| + | [[Image:k1378001-3.png|400px|center|Characterization of popular BioBrick RBSs]] | ||
| + | <p>1. By adding the protein that we have designed, and letting it react with the MC-LR, we find that they successfully reacted just as we expected.</p> | ||
| + | <p>2. From the HPLC results, we find that as the ratio of the mlrA we have designed increases, the percentage of the reaction product increases and the amount of MC-LR decreases consequencely. This result successful indicates that the protein mlrA we have designed is able to react with MC-LR and release the product which is 160 times less toxic than MC-LR, as we mentioned in the Background.</p> | ||
| + | |||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
===Functional Parameters=== | ===Functional Parameters=== | ||
<partinfo>BBa_K1378001 parameters</partinfo> | <partinfo>BBa_K1378001 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| + | <span class='h3bb'>Sequence and Features</span> | ||
| + | <partinfo>BBa_K1378001 SequenceAndFeatures</partinfo> | ||
Revision as of 13:15, 19 October 2019
MlrA

Introduction
MlrA is a 28kDa protease found in Sphingomonas sp. which can cleavage microcystins(MCs).
MlrA is one part of the gene cluster responsible for the ability of MC degradation. The cluster includes four ORFs, mlrA, mlrB, mlrC and mlrD, which can hydrolyze MCs and facilitate absorption of the products as carbon source. MlrA is sometimes referred as a metalprotease by inhibitor studies.
MlrA can cleavage the Adda-Arg bond and causes ring opening.(Fig. 1) The first-step linearized product shows much weaker hepatoxin compared with MCs. In the experiment of mouse bioassay, up to 250 mg/kg of linearized MC-LR shows no toxicity to mouse, much higher than 50% lethal dose 50mg/kg of cyclic MC-LR. Furthermore, the linearization also raise the median inhibition concentration to 95nM, around 160 times higher than original 0.6nM. [1]

Usage and Biology
Characterization
1. Constructing method for analysis of MC concentration
p-Nitrophenyl phosphate (pNPP) is a widely used non-specific substrate to test protein phosphatase activity and it can be hydrolyzed to p-Nitrophenyl(pNP) with characteristic absorption at 405nm. The measurement of PP1 activity is based on the accumulation of pNP. Considering the microcystin (MC) is the inhibitor of PP1 and MlrA can disrupt MC’s structure to disrupt its inhibitory effect, the MlrA activity can be detected by quantification of absorption at 405nm (Fig. 2). So the concentration of MCs after degradation can be finally measured by absorption spectrophotometry method with all the calibration curves for all the interactions above.

Firstly a calibration curve of PP1 activity was generated. The concentration of substrate pNP is sufficient overall so the PP1 enzyme is saturated and proportion to the accumulation rate of product pNPP. We could select a proper working concentration of PP1 in the range of nearly linear relationship between PP1 and change rate of 405nm absorption.

Based on the premise of linear relationship between product and absorbance, we choose 0.05unit/ul as the working concentration of PP1 and then test the inhibition efficiency of MC-LR. As a result, PP1 activity decreases after the addition of MC-LR and there is a positive correlation between the reduction of absorbance and concentration of MC-LR.

2. Verifying the degradation effect of MlrA
To test the degradation efficiency of MlrA expressed by E. coli, MlrA expression plasmid has been constructed and transformed into E. coli strain BL21(DE3) (Fig. 5a). After induction, the bacteria are lysed by lysozyme and incubated with MC solution. Judged by PP1 activity treated by the mixture, the activity in experiment group expressing MlrA is much higher than strain carrying blank vectors, suggesting that MC-LR is degraded (Fig. 5b). Therefore, it could be concluded that MlrA function well in E. coli expression system.

References
[1] Gehringer, M. M., Milne, P., Lucietto, F., & Downing, T. G. (2005). Comparison of the structure of key variants of microcystin to vasopressin. Environmental toxicology and pharmacology, 19(2), 297-303.
Contribution
Aalto-Helsinki 2016 team has contributed to this part by improving its sequence and characterization. The part number is BBa_K1907002.
SBS_SH_112144 2018 team has improved parts mlrA gene BBa_K1378001 by modifying its sequence, adding 6x His tag, as well as making it a segment of composite parts. Its part number is BBa_K2888010.
shanghai-HS 2019's characterization
BBa_K1378001 MlrA
Methods:
1. replicating the MlrA DNA through a technology called PCR, Gel Electrophoresis and Extraction of MlrA
2. combining the gene MlrA with plasmids, and transformed the plasmids to DH5-alpha, a strain of E. coli.
3. Senting the extracted plasmid to Sangon, a biotech company, for DNA sequencing after a series of procedures.
4. transforming the plasmid to E. coli BL21(DE3), another type of E-coli characterized in producing protein, to express the targeted protein.
5. purifing the protein MlrA and utilizing HPLC to determine the effectiveness of the protein MlrA.
Results:
By adding the protein that binds to the Ni21-NTA to the affinity column, we were able to collect the liquid that went through. We ran the SDS-Page Gel with thirteen chosen samples to verify the results of the protein purification. It turns out, we successfully purified pET28-mlrA1, indiciating that it can be devoted to degrading MC-LR.
Results:
Protein Purification
As you can see from the picture, we use N-Dodecyl-β-D-maltoside, which is also known as DDM, to transfer the target membrane protein mlrA from the supernatant to the membrane of DDM. Since DDM provides the membrane which mlrA depends on, with the DDM concentration increases, the concentration of the mlrA in the supernatant decreases to zero. That is the last step of protein purification.
After centrifuge the solution, to further purify the protein, we put Ni21-NTA with the supernatant and incubate overnight. The mlrA will be combined with the specific binding sites on Ni21-NTA. Then, using the method of gradient elution, while the amount of imidazole, which is a competitive agent in this step, increases, it will compete with mlrA on the binding sites of the Ni column. Therefore, the mlrA is gradually elated.
Enzyme Function Test
High Performance Liquid Chromatography (HPLC)
To quantify whether MC-LR can be degraded from the samples, we conducted a control variable experiments, and the following are the data we got:
1. By adding the protein that we have designed, and letting it react with the MC-LR, we find that they successfully reacted just as we expected.
2. From the HPLC results, we find that as the ratio of the mlrA we have designed increases, the percentage of the reaction product increases and the amount of MC-LR decreases consequencely. This result successful indicates that the protein mlrA we have designed is able to react with MC-LR and release the product which is 160 times less toxic than MC-LR, as we mentioned in the Background.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 244
Illegal AgeI site found at 373 - 1000COMPATIBLE WITH RFC[1000]