Difference between revisions of "Part:BBa K3147010"
(→I. Part BBa_K3147010 (MBP-TEVcs-TEVPE10-VHH) function) |
(→II. Proof of function) |
||
| Line 10: | Line 10: | ||
===II. Proof of function=== | ===II. Proof of function=== | ||
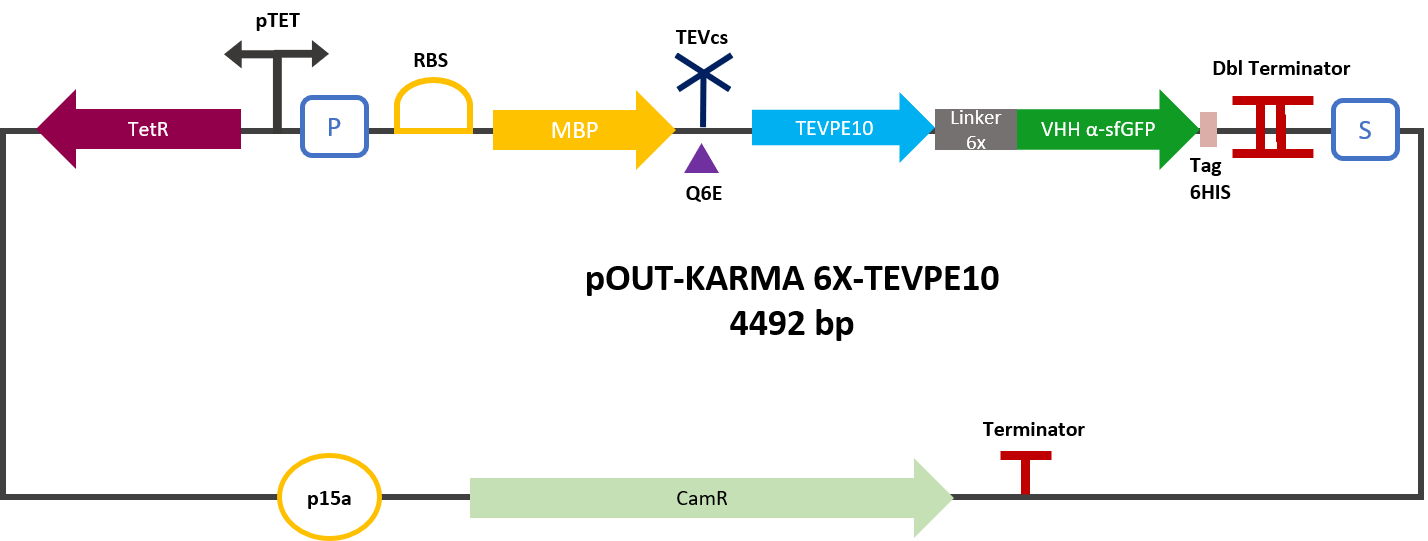
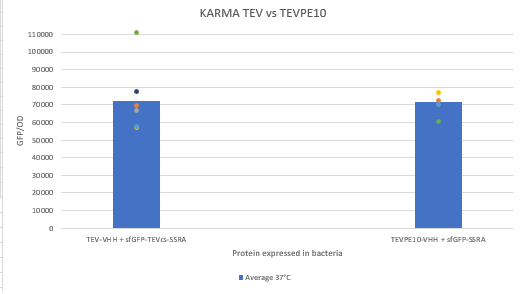
| − | The construction was cloned by Gibson Assembly in a pOUT18 vector under the control of a Tet-on promoter in order to control its expression. The experimental approach to test protease activity is to compare the fluorescence | + | The construction was cloned by Gibson Assembly in a pOUT18 vector under the control of a Tet-on promoter in order to control its expression. The experimental approach to test protease activity is to compare the fluorescence recovery of sfGFP against MBP-TEV-VHH and MBP-TEVPE10-VHH. In this experiment, basal controls of maximum and minimum fluorescence of reporter genes were used. Fluorescence data are obtained by plate reader and are expressed in arbitrary units relative to optical density at 600 nm. The protease is expressed by inducing the Tet promoter with 50 ng/mL of aTc (anhydrotetracycline). |
<div align="center">[[File:plasmideK3147010.png|450px]]</div> | <div align="center">[[File:plasmideK3147010.png|450px]]</div> | ||
Revision as of 15:31, 18 October 2019
I. Part BBa_K3147010 (MBP-TEVcs-TEVPE10-VHH) function
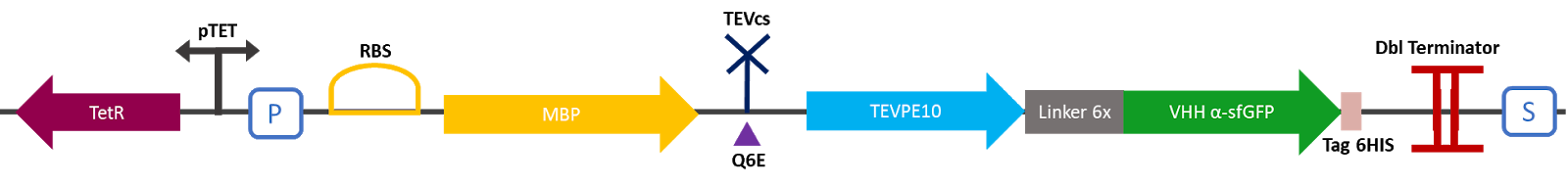
The 2019 Montpellier iGEM team made this construction in order to compare the basal activity of the TEV PE10 protease with the same protease fused to a VHH nanobody. This construction produces an MBP fused to a TEV protease attached to a VHH specific for sfGFP. A modified TEV cutting site was added between the MBP and the protease. MBP increases the solubility of the fusion protein [1], preventing the aggregation of the protein of interest. This stabilizes the expression and the sequence of the produced MBP does not have a signal peptide, which allows the protein to remain in the cytosol. The TEV cutting site allows self-cleavage of MBP from TEV once the protein is produced [2]. A 6His affinity tag was added to C-terminus of the VHH to facilitate validation of protein production and MBP cleavage via Western blot and protein purification if desired.
II. Proof of function
The construction was cloned by Gibson Assembly in a pOUT18 vector under the control of a Tet-on promoter in order to control its expression. The experimental approach to test protease activity is to compare the fluorescence recovery of sfGFP against MBP-TEV-VHH and MBP-TEVPE10-VHH. In this experiment, basal controls of maximum and minimum fluorescence of reporter genes were used. Fluorescence data are obtained by plate reader and are expressed in arbitrary units relative to optical density at 600 nm. The protease is expressed by inducing the Tet promoter with 50 ng/mL of aTc (anhydrotetracycline).
Reference
[1] Raran-Kurussi, Sreejith, et David S. Waugh. 2012. « The Ability to Enhance the Solubility of Its Fusion Partners Is an Intrinsic Property of Maltose-Binding Protein but Their Folding Is Either Spontaneous or Chaperone-Mediated » éd. Bostjan Kobe. PLoS ONE 7(11): e49589.
[2] Shih, Y.-P. 2005. « Self-Cleavage of Fusion Protein in Vivo Using TEV Protease to Yield Native Protein ». Protein Science 14(4): 936‑41.
[3] Yi, L. et al. 2013. « Engineering of TEV Protease Variants by Yeast ER Sequestration Screening (YESS) of Combinatorial Libraries ». Proceedings of the National Academy of Sciences 110(18): 7229‑34
[4] Kubala, M. H., Kovtun, O., Alexandrov, K., & Collins, B. M. (2010). Structural and thermodynamic analysis of the GFP:GFP-nanobody complex. Protein science : a publication of the Protein Society, 19(12), 2389–2401. doi:10.1002/pro.519
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 77
Illegal BsaI.rc site found at 2377
Illegal SapI.rc site found at 1868