Difference between revisions of "Part:BBa K584000"
| Line 8: | Line 8: | ||
The detailed qualitative and quantitative <u><b>[https://parts.igem.org/Part:BBa_I13453:Experience#User_Reviews review]</b></u> has been added into the experience page of the BBa_I13453 PBad promoter biobrick. | The detailed qualitative and quantitative <u><b>[https://parts.igem.org/Part:BBa_I13453:Experience#User_Reviews review]</b></u> has been added into the experience page of the BBa_I13453 PBad promoter biobrick. | ||
| − | |||
===Usage and Biology=== | ===Usage and Biology=== | ||
| + | |||
| + | <h2> <b> BOKU-Vienna 2019 - Characterization </b> </h2> | ||
| + | |||
| + | When designing our project, we planned on incorporating a viability signal that expresses GFP in case of living cells under the control of an arabinose inducible promoter (pBAD). Since the construct that we cloned contained an arabinose inducible promoter (BBa_I13453) that didn’t respond to induction despite correct sequence, we decided to test the respective composite part [[Part:BBa_K584000|BBa_K584000]] (pBAD + GFP generator) under different arabinose concentrations. <br> | ||
| + | The composite part [[Part:BBa_K584000|BBa_K584000]] was transformed into competent Escherischia coli (DH10B). Two clones were picked and grown on uninduced LB-media for 1 hour at 37°C on a shaker. 1mL aliquots were transferred into eprouvettes containing 1mL LB-media with different arabinose concentrations to an end concentration of 0, 0.0001, 0.001, 0.01, 0.05, 0.1 mM arabinose. Fluorescence was measured in a plate reader after 15 hours of incubation at 37°C on a shaker together with a fluorescein standard. OD-values were measured with a spectrophotometer right before samples were transferred into the 96-well plate.<br> | ||
| + | <br> | ||
| + | [[File:T--BOKU-Vienna--K584000FluorescenceAtDifArabCon.png|600px]] <br> | ||
| + | Figure 1: Fluorescence at different arabinose concentrations <br> | ||
| + | <br> | ||
| + | We calculated the mean fluorescence for each arabinose concentration across both clones, calculated the fluorescence for OD=1 and compared their fluorescence values (see figure 1). Fluorescence did not increase significantly after induction in comparison to the control of 0.04µM fluorescein, which equals the fluorescence of constantly expressed GFP under a medium strength promoter at OD<sub>600</sub>=0.5. <br> | ||
| + | <br> | ||
| + | To verify this data, the experiment was repeated as above, but with 8 hours incubation time and different arabinose concentrations (see figure 2). Again, as in the first experiment, '''no significant change in fluorescence could be measured after induction.''' <br> | ||
| + | <br> | ||
| + | [[File:T--BOKU-Vienna--K584000FluorescenceAtDifArabCon2.png|600px]] <br> | ||
| + | Figure 2: Fluorescence at different arabinose concentrations <br> | ||
| + | <br> | ||
| + | Since we could not observe an induction, we checked for correctness of the plasmid by restriction digestion with the enzymes EcoRI and PstI (see figure 3). <br> | ||
| + | [[File:T--BOKU-Vienna--K584000Restriction.png|300px]]<br> | ||
| + | Figure 3: Restriction digestion of BBa_K584000 - The band near 1kb matches the size of the insert, the band at 2kb the size of the empty pSB1C3 backbone <br> | ||
| + | <br> | ||
| + | The fact that we did not observe induction of gene expression by arabinose could have different reasons. The cells might lack transporters for arabinose (e.g. araE, araFGH) or are capable of effectively degrading arabinose (e.g. by expressing araBAD genes)(Jensen et al., 1993, https://doi.org/10.1111/j.1432-1033.1993.tb19885.x).<br> | ||
| + | <br> | ||
| + | Raw data from the plate reader [https://docs.google.com/spreadsheets/d/1afDS4MYlEaA8YAkA31RwLIPdH_yAG8frl5moVshdEF8/edit?usp=sharing here] <br> | ||
| + | <br> | ||
| + | Parameters: 485nm excitation, 525nm emission <br> | ||
| + | <br> | ||
| + | Materials: <br> | ||
| + | - LB-media with Chloramphenicol<br> | ||
| + | - Tecan Infinite 200 plate reader<br> | ||
| + | - white 96-well plates with white flat bottoms<br> | ||
| + | - Hitachi Photometer U-1900<br> | ||
| + | - L(+)-Arabinose (provider: ROTH; Art.-Nr. 5118.2)<br> | ||
| + | - 2% agarose gel<br> | ||
| + | - Quick-Load Purple 1kb Plus DNA Ladder | ||
| + | |||
<!-- --> | <!-- --> | ||
Revision as of 15:23, 18 October 2019
pBAD + GFP generator
Characterization
The [http://2011.igem.org/Team:KULeuven K.U.Leuven 2011 iGEM Team] has extensively characterized the existing Biobrick part PBad promoter(BBa_I13453).
The detailed qualitative and quantitative review has been added into the experience page of the BBa_I13453 PBad promoter biobrick.
Usage and Biology
BOKU-Vienna 2019 - Characterization
When designing our project, we planned on incorporating a viability signal that expresses GFP in case of living cells under the control of an arabinose inducible promoter (pBAD). Since the construct that we cloned contained an arabinose inducible promoter (BBa_I13453) that didn’t respond to induction despite correct sequence, we decided to test the respective composite part BBa_K584000 (pBAD + GFP generator) under different arabinose concentrations.
The composite part BBa_K584000 was transformed into competent Escherischia coli (DH10B). Two clones were picked and grown on uninduced LB-media for 1 hour at 37°C on a shaker. 1mL aliquots were transferred into eprouvettes containing 1mL LB-media with different arabinose concentrations to an end concentration of 0, 0.0001, 0.001, 0.01, 0.05, 0.1 mM arabinose. Fluorescence was measured in a plate reader after 15 hours of incubation at 37°C on a shaker together with a fluorescein standard. OD-values were measured with a spectrophotometer right before samples were transferred into the 96-well plate.

Figure 1: Fluorescence at different arabinose concentrations
We calculated the mean fluorescence for each arabinose concentration across both clones, calculated the fluorescence for OD=1 and compared their fluorescence values (see figure 1). Fluorescence did not increase significantly after induction in comparison to the control of 0.04µM fluorescein, which equals the fluorescence of constantly expressed GFP under a medium strength promoter at OD600=0.5.
To verify this data, the experiment was repeated as above, but with 8 hours incubation time and different arabinose concentrations (see figure 2). Again, as in the first experiment, no significant change in fluorescence could be measured after induction.
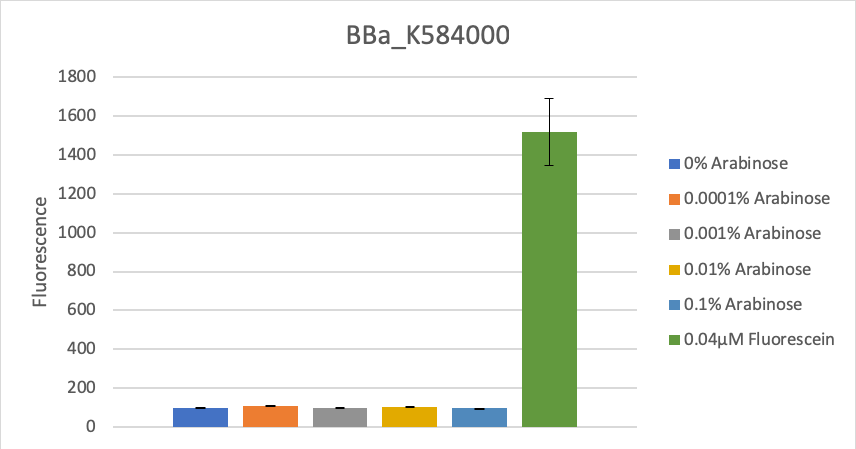
Figure 2: Fluorescence at different arabinose concentrations
Since we could not observe an induction, we checked for correctness of the plasmid by restriction digestion with the enzymes EcoRI and PstI (see figure 3).

Figure 3: Restriction digestion of BBa_K584000 - The band near 1kb matches the size of the insert, the band at 2kb the size of the empty pSB1C3 backbone
The fact that we did not observe induction of gene expression by arabinose could have different reasons. The cells might lack transporters for arabinose (e.g. araE, araFGH) or are capable of effectively degrading arabinose (e.g. by expressing araBAD genes)(Jensen et al., 1993, https://doi.org/10.1111/j.1432-1033.1993.tb19885.x).
Raw data from the plate reader here
Parameters: 485nm excitation, 525nm emission
Materials:
- LB-media with Chloramphenicol
- Tecan Infinite 200 plate reader
- white 96-well plates with white flat bottoms
- Hitachi Photometer U-1900
- L(+)-Arabinose (provider: ROTH; Art.-Nr. 5118.2)
- 2% agarose gel
- Quick-Load Purple 1kb Plus DNA Ladder
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 125
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 65
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 801
