Difference between revisions of "Part:BBa K515005"
Djcamenares (Talk | contribs) |
Djcamenares (Talk | contribs) (→Characterization for Universal Golden Gate Cloning (Alma College)) |
||
| Line 12: | Line 12: | ||
A depiction of the primer binding, and a representative gel, is below. | A depiction of the primer binding, and a representative gel, is below. | ||
| − | [[File:T--Alma--universalprimers2019_2.png]] | + | [[File:T--Alma--universalprimers2019_2.png|400px]] |
PCR products from some reaction combinations were selected and sequenced from the middle towards the end - this way, we can confirm if the overhang and BsaI site were added correctly. VF2 and VR primers were used as controls in the event that some primer pairs gave no or incorrect products. | PCR products from some reaction combinations were selected and sequenced from the middle towards the end - this way, we can confirm if the overhang and BsaI site were added correctly. VF2 and VR primers were used as controls in the event that some primer pairs gave no or incorrect products. | ||
| Line 18: | Line 18: | ||
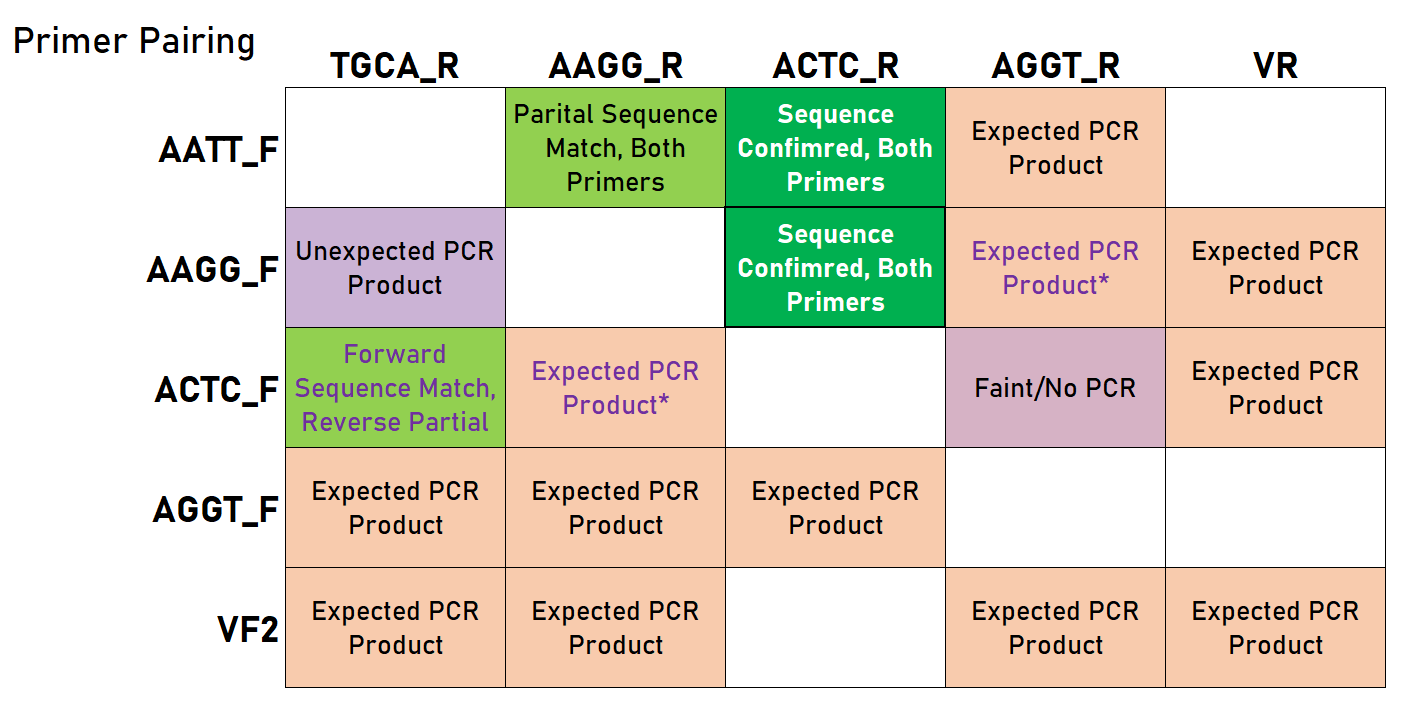
As you can see in the table below, we had mixed results - some of these PCRs required optimization, and were sensitive to annealing temperature and the speed at which the temperature shifted (evident by comparing the reaction in an old thermocycler versus a new mini8 PCR machine). However, we were able to show that this approach can be viable and does indeed lead to products that should work in a Golden Gate assembly reaction. | As you can see in the table below, we had mixed results - some of these PCRs required optimization, and were sensitive to annealing temperature and the speed at which the temperature shifted (evident by comparing the reaction in an old thermocycler versus a new mini8 PCR machine). However, we were able to show that this approach can be viable and does indeed lead to products that should work in a Golden Gate assembly reaction. | ||
| − | [[File:T--Alma--universalprimers2019_3.png]] | + | [[File:T--Alma--universalprimers2019_3.png|400px]] |
Legend: green indicates a successfully confirmed reaction (with the bold cell representing the result corresponding to the representative chromatogram above), light green indicates a sequencing analysis with partial success/confirmation/matching, orange-pink (EtBr color) indicates a PCR reaction that gave the correct product, and purple indicates a reaction that did not work. Empty cells represent untested combinations. *These reactions did not work initially, but were successful at least once (required further optimization). | Legend: green indicates a successfully confirmed reaction (with the bold cell representing the result corresponding to the representative chromatogram above), light green indicates a sequencing analysis with partial success/confirmation/matching, orange-pink (EtBr color) indicates a PCR reaction that gave the correct product, and purple indicates a reaction that did not work. Empty cells represent untested combinations. *These reactions did not work initially, but were successful at least once (required further optimization). | ||
Revision as of 03:07, 18 October 2019
superfolder GFP (sfGFP)
a translational unit of superfolder GFP
Characterization for Universal Golden Gate Cloning (Alma College)
We used this BioBrick to characterize a new approach to Golden Gate assembly - one inspired by a paper by Bryksin et al. In this approach, universal PCR primers add BsaI sites and overhang sequences to any BioBrick, thanks to a specialized mismatch priming. This eliminates the need to order separate primers for every BioBrick you want to assembly using Golden Gate. We designed forward primers with overhangs sequences AATT, AAGG, ACTC, AGGT, and reverse primers with overhangs AAGG, ACTC, AGGT, and TGCA. For more information, refer to the diagram to the right or visit the contribution page on our Wiki.
Using this (and other) BioBricks and the above primers, we carried out PCR reactions to determine if this approach was feasible. The reactions were generally setup as follows: 0.5uL of purified plasmid DNA for BioBrick BBa_K515005 was mixed with 9.5uL ddH20, 1.25uL of each primer (@ a 10uM concentration), and finally with 12.5uL of a 2X Mix for Q5 DNA Polymerase. Reaction conditions were setup as suggested by NEB for Q5 polymerase, typically with an annealing temperature between 51 and 55C. In some trials, 9uL of ddH20 and 0.5ul of DMSO were added.
A depiction of the primer binding, and a representative gel, is below.
PCR products from some reaction combinations were selected and sequenced from the middle towards the end - this way, we can confirm if the overhang and BsaI site were added correctly. VF2 and VR primers were used as controls in the event that some primer pairs gave no or incorrect products.
As you can see in the table below, we had mixed results - some of these PCRs required optimization, and were sensitive to annealing temperature and the speed at which the temperature shifted (evident by comparing the reaction in an old thermocycler versus a new mini8 PCR machine). However, we were able to show that this approach can be viable and does indeed lead to products that should work in a Golden Gate assembly reaction.
Legend: green indicates a successfully confirmed reaction (with the bold cell representing the result corresponding to the representative chromatogram above), light green indicates a sequencing analysis with partial success/confirmation/matching, orange-pink (EtBr color) indicates a PCR reaction that gave the correct product, and purple indicates a reaction that did not work. Empty cells represent untested combinations. *These reactions did not work initially, but were successful at least once (required further optimization).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 172
- 1000COMPATIBLE WITH RFC[1000]
This BioBrick has been sequence verified.
It is a component of the composite BBa_K515105