Difference between revisions of "Part:BBa K2912017"
m (→Usage and Characterization) |
(→Usage and Characterization) |
||
| Line 48: | Line 48: | ||
</div> | </div> | ||
| − | The R-bodies proteins were predicted to fully translate after 44 minutes. Click [https://2019.igem.org/Team:SZU-China/Model SZU-China 2019 Model_R-body] to see more. | + | The R-bodies proteins were predicted to fully translate after 44 minutes. Click [https://2019.igem.org/Team:SZU-China/Model SZU-China 2019 Model_R-body]to see more. |
SZU-China 2019 iGEM team has synthesized the R-bodies and taken the scanning electron microscope pictures of the R-bodies under pH=7 and pH=6 (Fig.5,6). | SZU-China 2019 iGEM team has synthesized the R-bodies and taken the scanning electron microscope pictures of the R-bodies under pH=7 and pH=6 (Fig.5,6). | ||
| Line 55: | Line 55: | ||
<center> | <center> | ||
<html><img src='https://2019.igem.org/wiki/images/d/d5/T--SZU-CHINA--R-body1.png' style="width:50%;margin:0 auto"> | <html><img src='https://2019.igem.org/wiki/images/d/d5/T--SZU-CHINA--R-body1.png' style="width:50%;margin:0 auto"> | ||
| − | |||
<center> Fig.5 The Regulation of pH on R-Bodies </center></html></center> | <center> Fig.5 The Regulation of pH on R-Bodies </center></html></center> | ||
</div> | </div> | ||
Revision as of 20:42, 16 October 2019
Refractile inclusion bodies_ Caedibacter
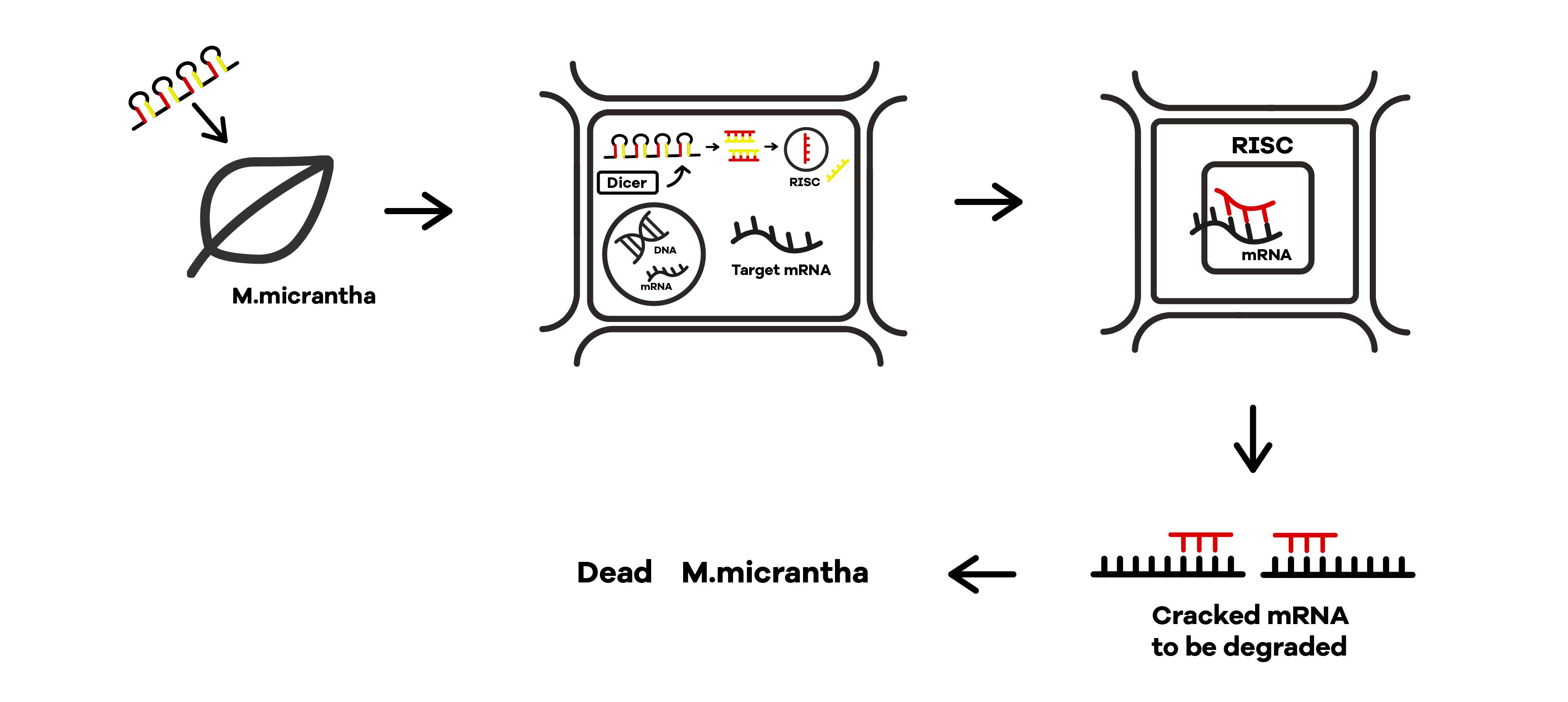
Refractile inclusion bodies SZU-China 2019 iGEM team was going to find a suicide switch inside the E coli that can break the whole body of the bacteria leading to the release of RNAi molecules transcribed from E coli inducing by IPTG or some other else. Therefore, we were in need the useful mechanism. Fortunately, we finally found the Refractile inclusion bodies (R-bodies) to kill the E coli, causing the inclusion to flow out of the plasma membrane, so that we can get the RNAi molecules transcribed by E coli. Refractile inclusion bodies, known as R bodies, are produced by only a few species of bacteria. These inclusion bodies are highly insoluble protein ribbons, typically seen coiled into cylindrical structures within the cell[1]. R-bodies are produced by Paramecium endosymbionts belonging to the genus Caedibacter. These intracellular bacteria confer upon their hosts a phenomenon called the killer trait[2]. This is one of the DNA sequences for the R body locus (reb) from Caedibacter taeniospiralis. The R bodies of C. taeniospiralis are type 51. They are about 0.5 μm wide, have a maximum length of 20 μm, and 13 nm thick, possess acute angles at each end, and unroll in a telescopic fashion when exposed to a pH of 6.5 or lower. These proteinaceous ribbons are rolling up inside the cell to form a hollow cylinder about 0.5 μm in diameter and 0.5 μm long. Sequence and Features
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 15
Illegal NheI site found at 38 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
SZU-China 2019 iGEM team
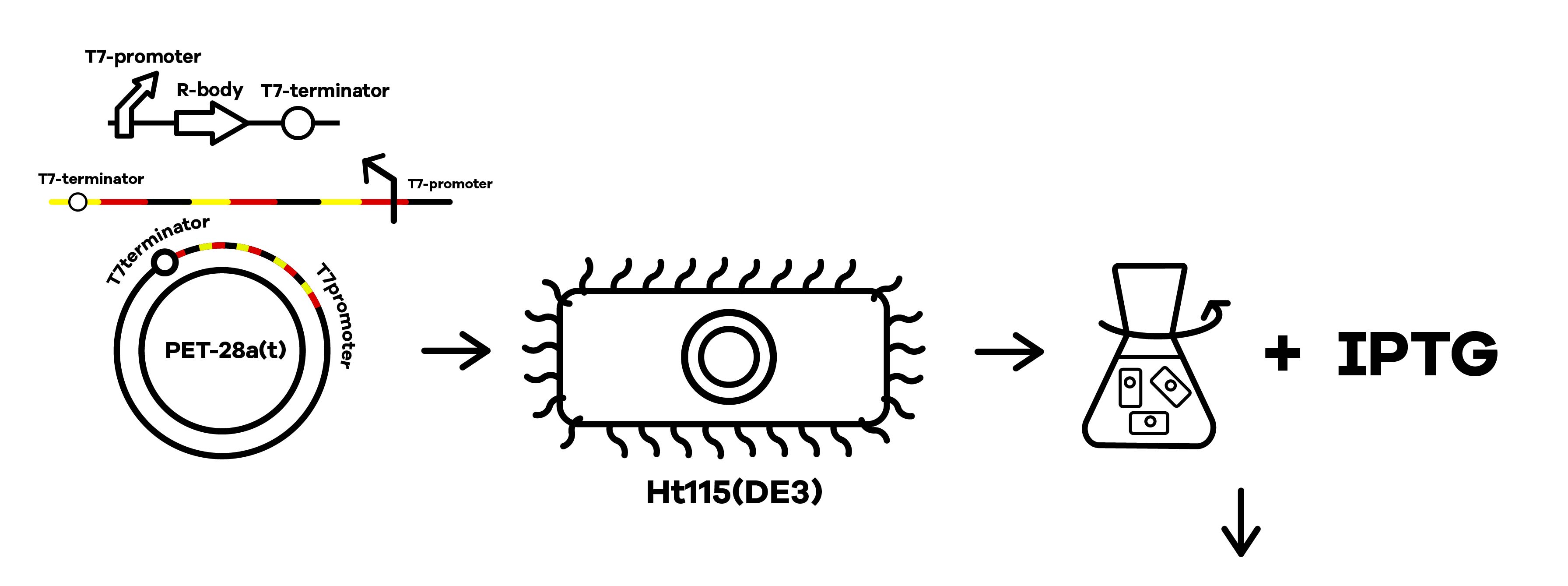
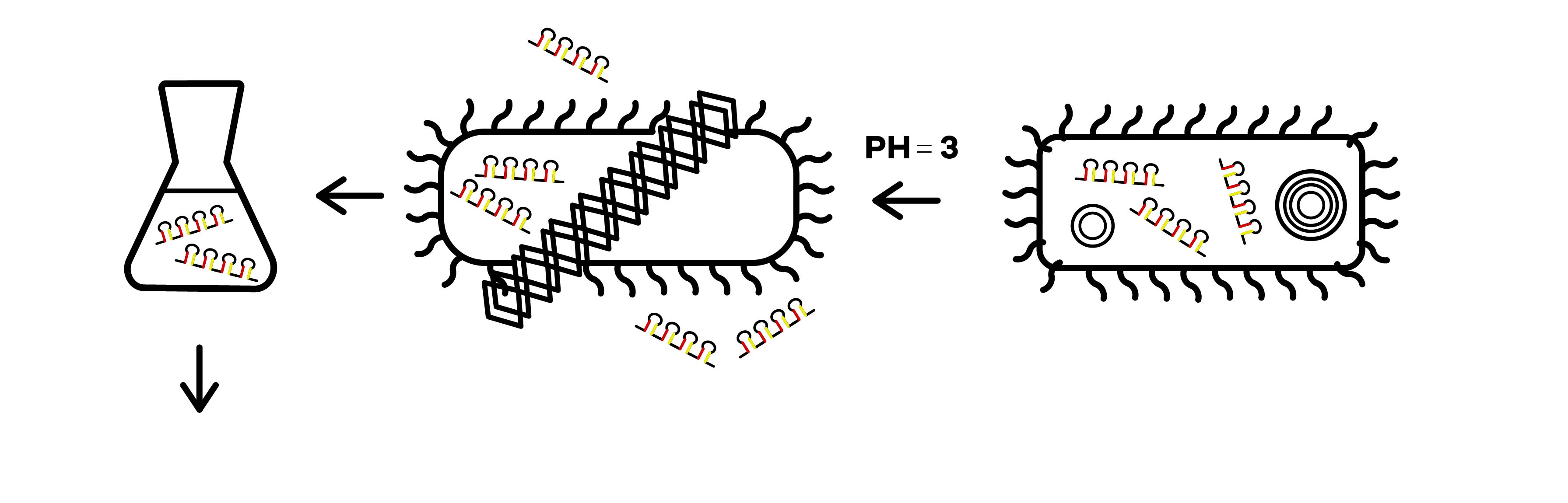
SZU-China 2019 iGEM team was going to find a suicide switch inside the E. coli that can break the whole body of the bacteria leading to the release of RNAi molecules transcribed from E. coli inducing by IPTG or some other else. Therefore, we required a useful mechanism. Fortunately, we finally found the ’’’Refractile inclusion bodies (R-bodies)’’’ to kill the E. coli, causing the inclusion to flow out of the plasma membrane so that we can get the RNAi molecules transcribed by E. coli (Fig.1).
Refractile inclusion bodies
Refractile inclusion bodies, known as R bodies, are produced by only a few species of bacteria. These inclusion bodies are highly insoluble protein ribbons, typically seen coiled into cylindrical structures within the cell[1]. R-bodies are produced by Paramecium endosymbionts belonging to the genus Caedibacter. These intracellular bacteria confer upon their hosts, a phenomenon called the killer trait[2]. The R bodies of ’’C. taeniospiralis’’ are type 51. They are about ’’’0.5 μm wide’’’, have ’’’a maximum length of 20 μm’’’, and 13 nm thick, possess acute angles at each end and ’’’unroll’’’ in a telescopic fashion when ’’’exposed to a pH of 6.5 or lower’’’. These proteinaceous ribbons are rolling up inside the cell to form ’’’a hollow cylinder’’’ about ’’’0.5 μm in diameter’’’ and 0.5 μm long[3].
Usage and Characterization
SZU-China 2019 iGEM team constructed a plasmid with BBa_K1475900 (Promoter),BBa_K2912014 (Attenuator),BBa_K2912000 (Reb A),BBa_K2912001 (Reb B),BBa_K2912003 (Reb D),BBa_K2912002 (Reb C),BBa_B0010 (Terminator)(Fig.2).
The attenuator was designed to control the expression of R-body via the concentration of Tryptophan. When the concentration of Trp is under 0.3%, R-body begins to express. Hence, after our RNAi molecules are adequately transcribed inside E. coli, the R-body proteins can be induced to translate to form the R-bodies rolled-up inside the bacteria. Then, we will change the pH of the media to induce the R-bodies to unroll and crack the E. coli after the R-bodies are fully expressed.
The R-bodies proteins were predicted to fully translate after 44 minutes. Click SZU-China 2019 Model_R-bodyto see more.
SZU-China 2019 iGEM team has synthesized the R-bodies and taken the scanning electron microscope pictures of the R-bodies under pH=7 and pH=6 (Fig.5,6).

References
[1] Pond FR, Gibson I, Lalucat J, et al. R-body-producing Bacteria.[J]. Microbiol Rev, 1989, 53(1): 25-67.
[2] Matsuoka JI, Ishizuna F, Kurumisawa K, et al. Stringent Expression Control of Pathogenic R-body Production in Legume Symbiontazorhizobium Caulinodans[J]. Mbio, 2017, 8(4): 0-17.
[3] Heruth DP, Pond FR, Dilts JA, et al. Characterization of Genetic Determinants for R Body Synthesis and Assembly in Caedibacter Taeniospiralis 47 and 116.[J]. Journal of Bacteriology, 1994, 176(12): 3559-3567.
