Difference between revisions of "Part:BBa J23100"
Nickdelkis (Talk | contribs) |
|||
| Line 128: | Line 128: | ||
</body> | </body> | ||
</html> | </html> | ||
| + | |||
| + | |||
| + | ===iGEM Team 2019 Stuttgart Characterization=== | ||
Revision as of 14:26, 16 October 2019
constitutive promoter family member . For video explanation on this promoter family and its use, see [http://www.synbiotrails.org/#!/trail?id=org.clothocad.trails.youtube.TranscriptionModels this].
Variant RFP (au) J23112 1 J23103 17 J23113 21 J23109 106 J23117 162 J23114 256 J23115 387 J23116 396 J23105 623 J23110 844 J23107 908 J23106 1185 J23108 1303 J23118 1429 J23111 1487 J23101 1791 J23104 1831 J23102 2179 J23100 2547 |
Constitutive promoter family
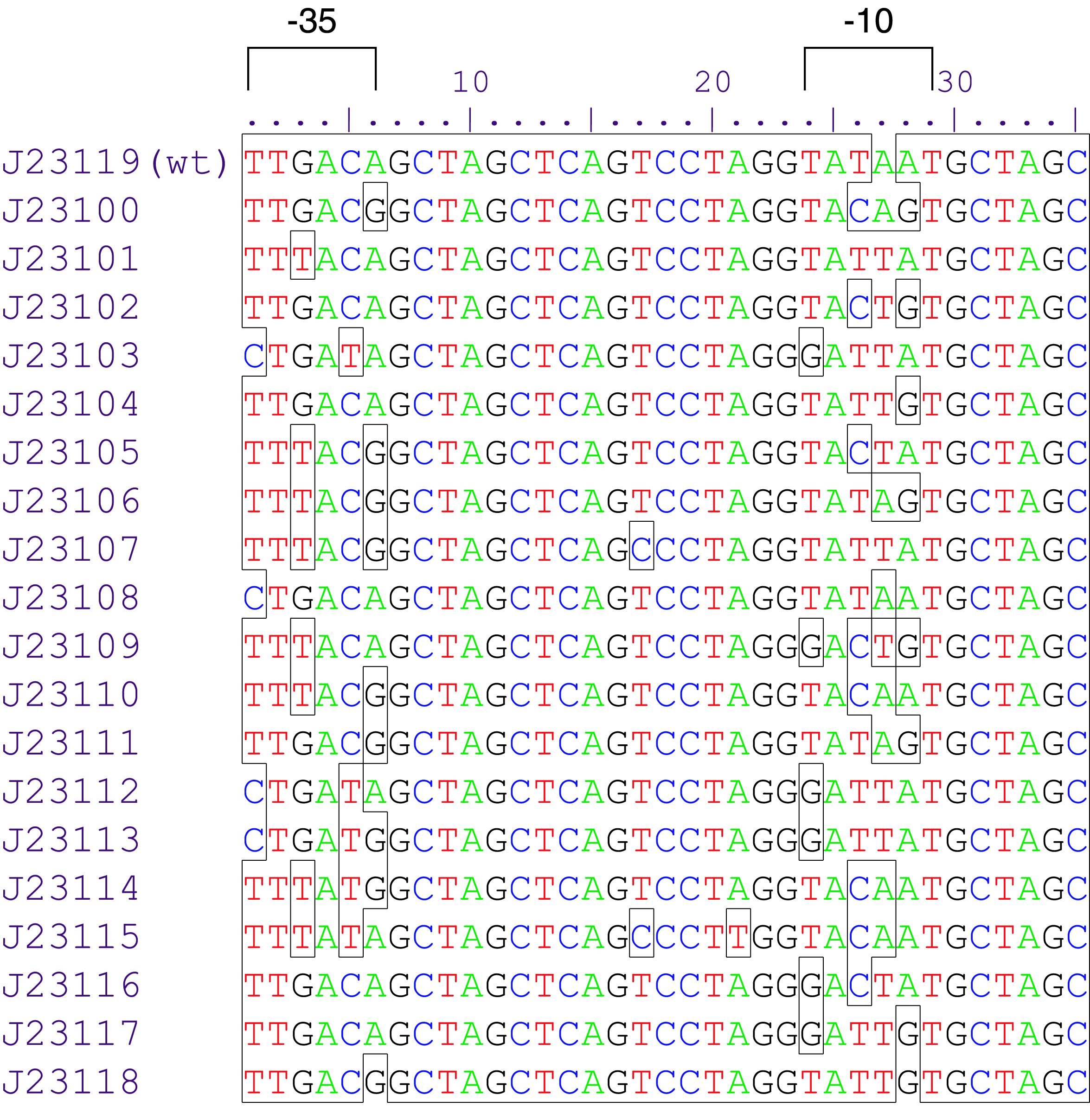
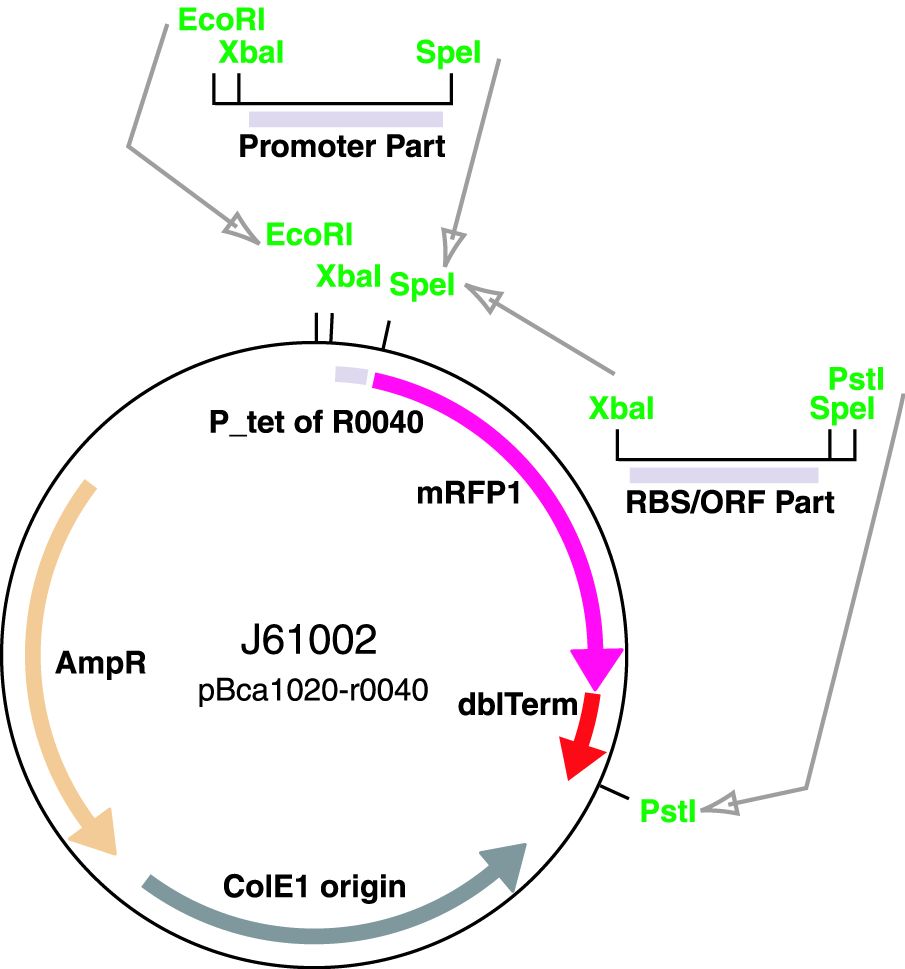
Parts J23100 through J23119 are a family of constitutive promoter parts isolated from a small combinatorial library. J23119 is the "consensus" promoter sequence and the strongest member of the family. All parts except J23119 are present in plasmid J61002. Part J23119 is present in pSB1A2. This places the RFP downstream of the promoter. Reported activities of the promoters are given as the relative fluorescence of these plasmids in strain TG1 grown in LB media to saturation. See part BBa_J61002 for details on their use.
These promoter parts can be used to tune the expression level of constitutively expressed parts. The NheI and AvrII restriction sites present within these promoter parts make them a scaffold for further modification. JCAraw
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Functional Parameters
Relative promoter strength estimates (see [http://2009.igem.org/Team:Groningen/Promoters this page] from Groningen 2009):
| Reference | Strength |
|---|---|
| BBa_J23109 | 67 |
| BBa_J23106 | 2 |
USTC_2009's MEAUREMENT
[K176011]
Usage and Biology
The Yale iGEM team has assembled this promoter to citrine (an improved version of YFP, with excitation peak at 514nm and emission peak at 527nm) and a T7-terminator to quantify the level of expression in E. coli and in non-model organism hosts. This construct is available as the biobrick K185000
This construct has been successfully cloned into E. coli using the broad-host range vector pKT230, a RSF1010 derived plasmid, as well as using the pPZP200 plasmid which can be transformed into agrobacterium and rhizobium. Leaky expression of citrine was observed.
Yale 2015's Characterization
Measured strength
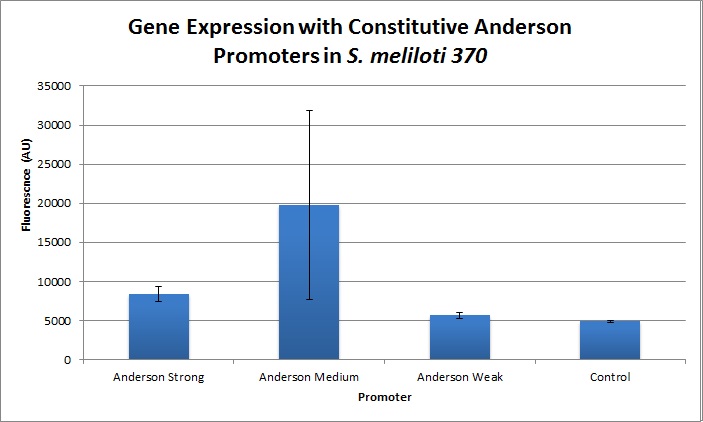
The Yale iGEM team found fluorescence levels nine times above the baseline level in E. coli cells transformed with the promoter-citrine construct. We also found this promoter to be effective in driving gene expression in S. meliloti, with fluorescence levels of over two times that of the baseline observed.
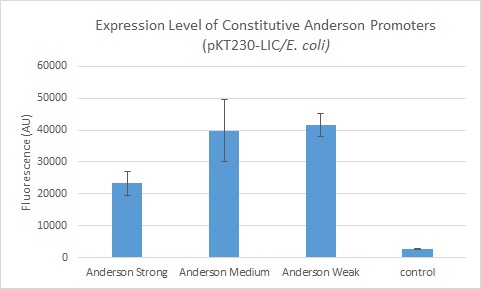
We further compared expression level of this promoter to two of the other promoters in the Anderson collection. The Anderson lab found relatively expression levels of 1, 0.33, and 0.10 respectively when comparing the promoters J23100 (Anderson Strong), J23110 (Anderson Medium) and J23114 (Anderson Weak). We found the relative expressions to be 1, 1.71 and 1.78 when we transformed our constructs into E.coli: K1856004 (Anderson Strong-Citrine), K1856003 (Anderson Medium-Citrine), K1856002 (Anderson Weak-Citrine). This highlights that even as all three Anderson promoters are effective in driving gene expression in E. coli, the relative expression level may vary depending on the gene fused to the promoter and on the E. coli strain used. The Yale iGEM team's readings were obtained in E. coli DH10B.
Comparing expression levels in S. meliloti (a rhizobium strain), the relative expression values were 1, 2.36, and no significant expression for the Anderson Strong, Anderson Medium, and Anderson Weak promoters respectively.
Team Warsaw 2010's measurement
Absolute promoter strength: 13,1pg RNA/minute/ug substrate DNA. It equals 2,80 microPoPSSheffield 2016's Characterisation
Nanjing-China 2017's Characterisation
In order to further extend the range and improve the sensitivity of detection, we expect to enhance the expression of SQR by substituting psqrR with pcons, the most efficient promotor among the igem parts. By doing so, we make sure that there is enough SQR in the system to oxidize even a small amount of S2- in time.
We have observed RFP response after inducing the modified pathway with 100 μmol/L Na2S, which proves that the new pathway works. We are still working on the comparison of two pathways.
>Internal Priming Screening Characterization of BBa_J23100: Has no possible internal priming sites between this BioBrick part and the VF2 or the VR primer.
The 2018 Hawaii iGEM team evaluated the 40 most frequently used BioBricks and ran them through an internal priming screening process that we developed using the BLAST program tool. Out of the 40 BioBricks we evaluated, 10 of them showed possible internal priming of either the VF2 or VR primers and sometime even both. The data set has a range of sequence lengths from as small as 12 bases to as large as 1,210 bases. We experienced the issue of possible internal priming during the sequence verification process of our own BBa_K2574001 BioBrick and in the cloning process to express the part as a fusion protein. BBa_K2574001 is a composite part containing a VLP forming Gag protein sequence attached to a frequently used RFP part (BBa_E1010). We conducted a PCR amplification of the Gag-RFP insert using the VF2 and VR primers on the ligation product (pSB1C3 ligated to the Gag + RFP). This amplicon would serve as template for another PCR where we would add the NcoI and BamHI restriction enzyme sites through new primers for ligation into pET14b and subsequent induced expression. Despite gel confirming a rather large, approximately 2.1 kb insert band, our sequencing results with the VR primer and BamHI RFP reverse primer gave mixed results. Both should have displayed the end of the RFP, but the VR primer revealed the end of the Gag. Analysis of the VR primer on the Gag-RFP sequence revealed several sites where the VR primer could have annealed with ~9 - 12 bp of complementarity. Internal priming of forward and reverse primers can be detrimental to an iGEM project because you can never be sure if the desired construct was correctly inserted into the BioBrick plasmid without a successful sequence verification.
Measured strength
Sheffield 2016 has improved the characterisation of both BBa_J23100 and BBa_J23106. These parts are a strong and medium promoter respectively, that we have used to design our iron detecting device. We have experimentally validated through fluorimetry that there is indeed a significant difference between expression levels of GFP coupled to the strong and medium promoters. Comparative analysis of promoter strengths can be directly interpreted from the data we obtained. This data can be found both on the original part experience pages of BBa_J23100 and BBa_J23106, as well as at our website. Our team has also improved the function of the BBa_J23100 part, by adding an RBS site downstream of the promoter (BBa_K2016005). BBa_J23100 was originally submitted to the Registry by Berkeley 2006.

Fluorescence of JC28 mutants or W3110 wild types transformed with RyhB-GFP constructs under the control of medium (MedGFP) or strong promoters (StrGFP).
Improvement from iGEM 2018 UESTC-China
This year, we UESTC-China have improved the previous part BBa_J23100 by adding RBS and pelB-5D. The combination of promoter, RBS and signal peptide makes it convenient to use as a biobrick and gives it the function of extracellular expression. And the part number is BBa_K2617017. For the function determination, we chose the PETase as our reporter protein because its activity can be easily detected. Thus we constructed two plasmids with and without pelB-5D respectively to compare the function of our improved part and previous part.
Link to our biobrick: https://parts.igem.org/Part:BBa_K2617017.
The activity of the extracelluar fractions was detected by the method of pNPB. And the results were as followed(Fig. 1).
Based on our functional determination test results, the improvement that we did has succeeded. PETase that carrying our improved part (Fig. 1) has much higher activity level than the PETase that carrying promoter BBa_J23100 but without pelB-5D. From our experiment result, we could conclude that our improved part indeed gives the promoter BBa_J23100 the function of extracellular expression.
Thessaly 2019 Characterization
Thessaly 2019 sought to characterize the coding sequence of TEM-optimized beta-lactamase (BBa_I757010) under the regulation of the constituve Anderson Family promoters BBa_J23100, BBa_J23105, BBa_J23106, BBa_J23119. Beta-lactamase is an enzyme that hydrolyses beta-lactams (e.g. ampicillin) and is naturally found in procaryotic cells. A colorimetric assay has been developed using nitrocefin as a substrate which after hydrolysis from beta-lactamase changes the reaction color, from yellow (380nm) to red (490nm).
To achieve that, the coding sequence was assembled with each promoter, a universal RBS (BBa_B0034) and a double terminator(BBa_B0015). The parts were cloned in pSB1C3 and pSB1K3 and transformed into E. coli DH5a competent cells. For protein expression, the plasmids were transformed into E. coli BL21 (DE3) competent cells.
For the beta-lactamase assay, we set up the following experimental design:
1. Grow BL21 (DE3) cells overnight in 5ml LB (~16h) at a shaken incubator, 37 degrees C / 210rpm
2. The following morning, measure the OD600 of overnight cultures
3. Dilute all cultures to OD600¬ = 0.05 in M9 minimal medium
4. Grow cells 37 degrees C /210 RPM until OD600=0.4-0.6 (~2h)
5. Dilute all cells to the same OD600 (e.g. 0.4)
6. Load 160 of culture in a 96-well plate (do triplicates). Add 40 ul 0.5 uM nitrocefin for a final concentration of 100nM
7. Measure the absorbance at 490nm (for nitrocefin hydrolysis) and 600nm (for cell growth) every 30 seconds for 25 minutes in a microplate reader. Shake between measurements.
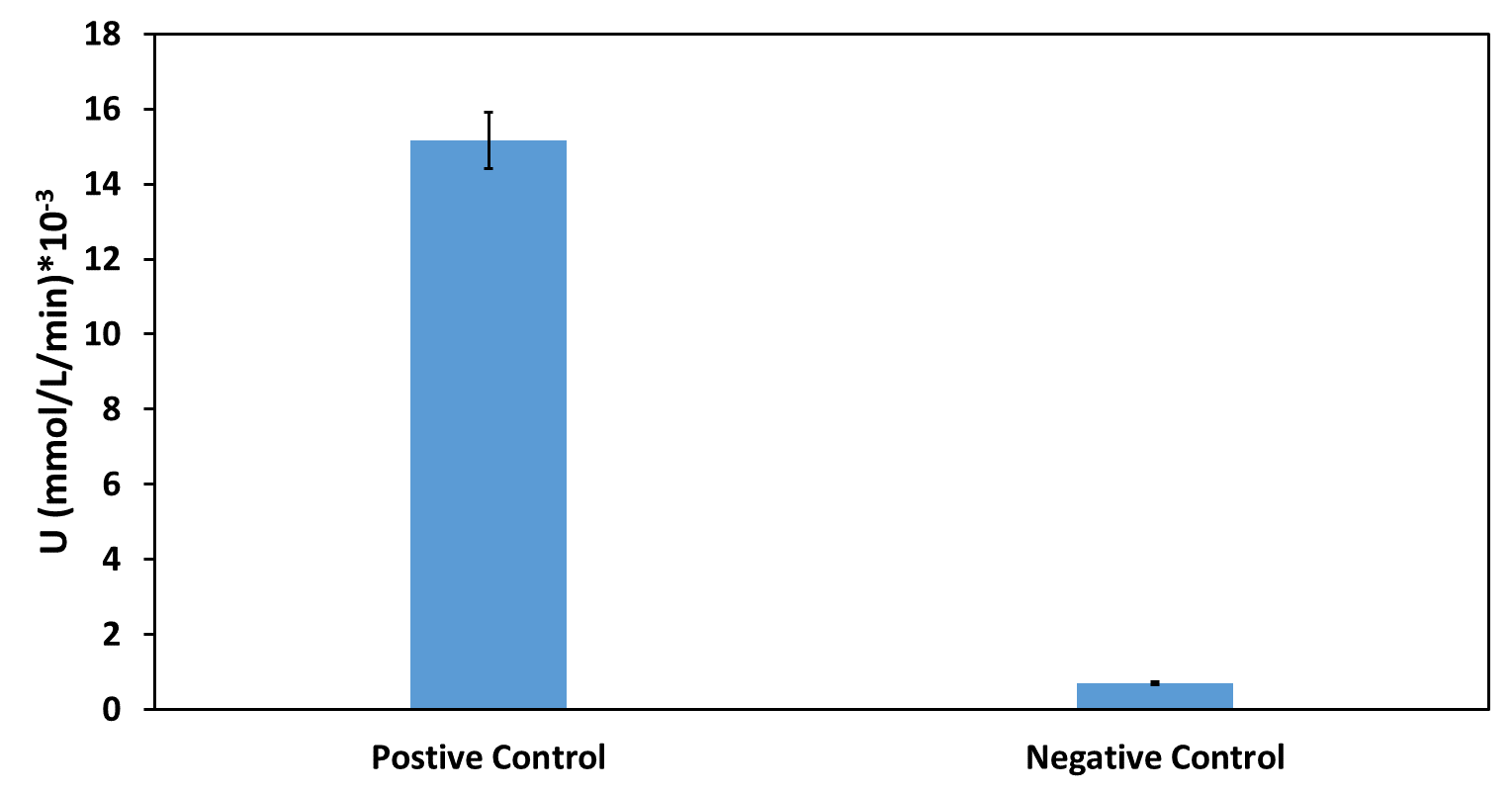
To ensure that the absorbance shown corresponds only to enzymatic activity by beta-lactamase, we included 3 controls in the experiment. The first control has M9 medium only (no cells) and nitrocefin, the second has empty BL21 (DE3) cells (no plasmid) and nitrocefin, while the third has BL21 (DE3) cells containing the plasmid but not the part (empty plasmid). To obtain comparable results, we normalized all values by dividing OD490 by OD600.
The results are shown in the graph below