Difference between revisions of "Part:BBa K3128001"
(→References) |
(→Usage and Biology) |
||
| Line 49: | Line 49: | ||
<div style="text-align:justify;"> | <div style="text-align:justify;"> | ||
| − | + | '''[https://2019.igem.org/Team:Grenoble-Alpes NeurDrop]''' detection system is based on the use of a '''BACTH'''. The point is to allow the induction of the gene only when the two sub-parts of AC are physically close, which only occurs when the target is present in the sample. The reassembly of AC enables cAMP production, followed by the '''transcription of the reporter''' that is under a '''CAP-cAMP dependent promoter'''. <br> | |
| + | To use this system, an '''AC deficient bacteria strain''' (BTH101) that is not able to produce cAMP endogenously is needed to prevent any transcription from CAP dependant promoter if there is no interaction between both sub-parts. <br> | ||
| + | For the promoter, we decide to use the '''lactose promoter''' (a CAP dependent promoter), we have demonstrated its repression in the absence of an exogenous source of cAMP (in the AC deficient bacterial strain). | ||
Revision as of 19:09, 14 October 2019
NanoLuciferase reporter for BACTH assay
Contents
Sequence and features
A reporter plasmid was created to be used for BATCH assays. This reporter plasmid is composed of a cAMP inducible promoter (CAP dependant) driving the expression of a reporter gene.
We used a very well characterized iGEM part : BBa_J04450.
Characterization of this BioBrick done by Team Grenoble-Alpes 2019showed that the lactose promoter present in this BioBrick was not inducing gene expression of the following gene if there is no cAMP which makes this BioBrick useful for our system.
Construction
Restriction sites were added on both 5’ and 3’ ends of the Red Fluorescent Protein gene as such:

The mRFP1 was removed and replaced by the Nano Luciferase previously amplified from a commercial vector).
Nano Luciferase was chosen for its ability to produce luminescence without the need of ATP (ATP being the substrate of Adenylate Cyclase (AC)), it must be as available as possible and so not be used by our reporter protein) and for the high level of luminescence observed for a single protein .
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Usage and Biology
Bacterial Adenylate Cyclase Two-Hybrid (BACTH)
The principle lies on the interaction-mediated reconstitution of a signalling cascade in Escherichia coli. The messenger molecule involved in this cascade is the
cyclic adenosine monophosphate (cAMP) produced by the adenylate cyclase. Adenylate cyclase is an enzyme catalysing the cAMP production from ATP. It
physiologically participates to the cellular transmission.
This system involves the Bordetella pertussis adenylate cyclase which is the responsible agent for the pertussis disease.
Adenylate cyclase catalytic domain has the particularity to be splittable in two distinct parts: T18 and T25 sub-parts, unable to fonction unless they
reassociate. Each sub-part of the enzyme is fused with a protein of interest, either the bait or the prey protein chose beforehand by the experimentator.
If two proteins interact, then T18 and T25 are bring together and reconstitute a functional adenylate cyclase enzyme thus enabling cAMP production. Using
cya- bacteria – strain for whom the adenylate gene is deleted, involving an absence of this endogenous enzyme – a BACTH could be done with the creation of two
fusion proteins : the first one, fused at its N or C terminal intracellular end with the T18 sub-part; the second one fused with the T25 sub-part.
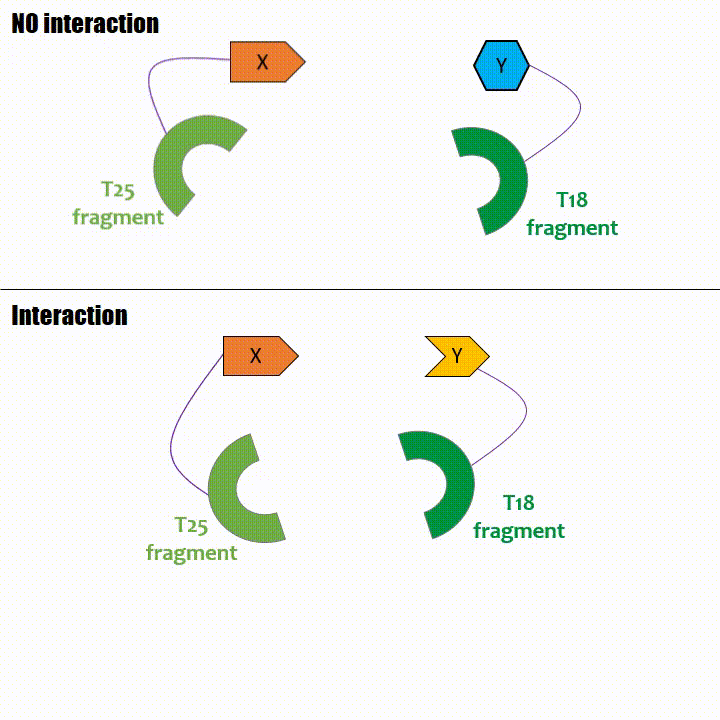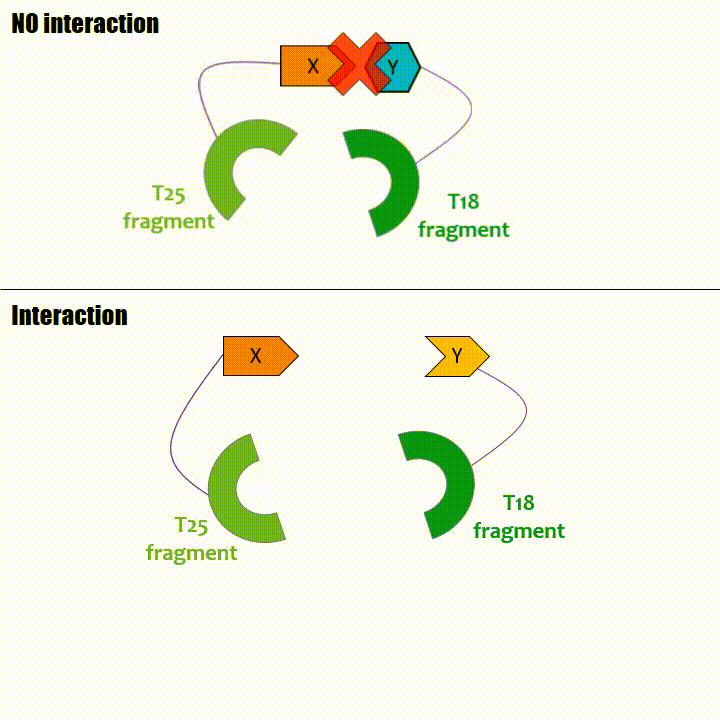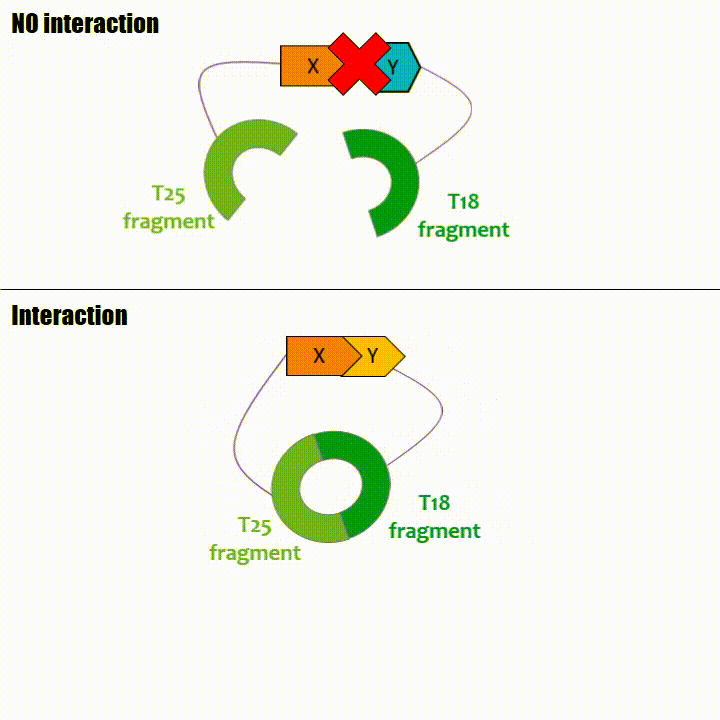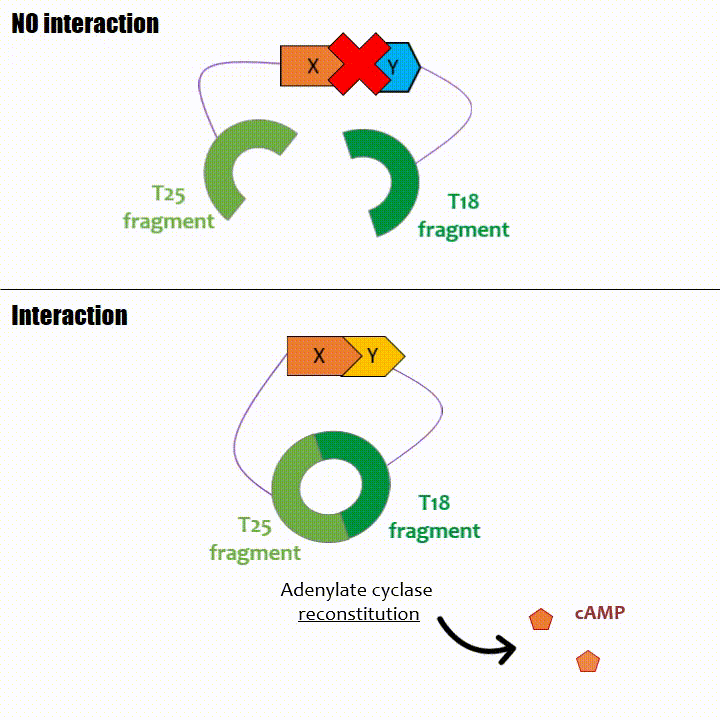
The interaction of these proteins of interest will lead to the adenylate cyclase reconstitution, thus initiating cAMP production. The cAMP produced will act as a
messenger by fixing itself to the transcriptional activator CAP, cAMP form the CAP-cAMP complex, controlling the expression of the lactose promoter by initiating transcription of the following gene.
This promoter is placed upstream the chosen reporter gene.
NeurDrop detection system is based on the use of a BACTH. The point is to allow the induction of the gene only when the two sub-parts of AC are physically close, which only occurs when the target is present in the sample. The reassembly of AC enables cAMP production, followed by the transcription of the reporter that is under a CAP-cAMP dependent promoter.
To use this system, an AC deficient bacteria strain (BTH101) that is not able to produce cAMP endogenously is needed to prevent any transcription from CAP dependant promoter if there is no interaction between both sub-parts.
For the promoter, we decide to use the lactose promoter (a CAP dependent promoter), we have demonstrated its repression in the absence of an exogenous source of cAMP (in the AC deficient bacterial strain).
References
Fields S, Song O. A novel genetic system to detect protein–protein interactions. Nature. (1989)
Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. PNAS. (1998)
Karimova G, Gauliard E, Davi M, P.Ouellette S, Ladant D. Protein–Protein Interaction: Bacterial Two-Hybrid. (2017)
Picture of the reaction ATP-cAMP. Khan Academy Website. Retrieved October 10, 2019, from https://www.khanacademy.org
Euromedex, BACTH System Kit
Leusch, Paulaitis, Friedman. Adenylate cyclase toxin of Bordetella pertussis: production, purification, and partial characterization. Am Soc Microbiol | Infect Immun. (1990)
Hantke, Winkler, Schultz. Escherichia coli exports cyclic AMP via TolC. J Bacteriol. (2011)
