Difference between revisions of "Part:BBa K1033933:Experience"
(→Applications of BBa_K1033933) |
(→Applications of BBa_K1033933) |
||
| Line 9: | Line 9: | ||
2019 iGEM team Linkoping Sweden validated this part.<br><br> | 2019 iGEM team Linkoping Sweden validated this part.<br><br> | ||
<b>Growth of p.cons-asPink</b><br> | <b>Growth of p.cons-asPink</b><br> | ||
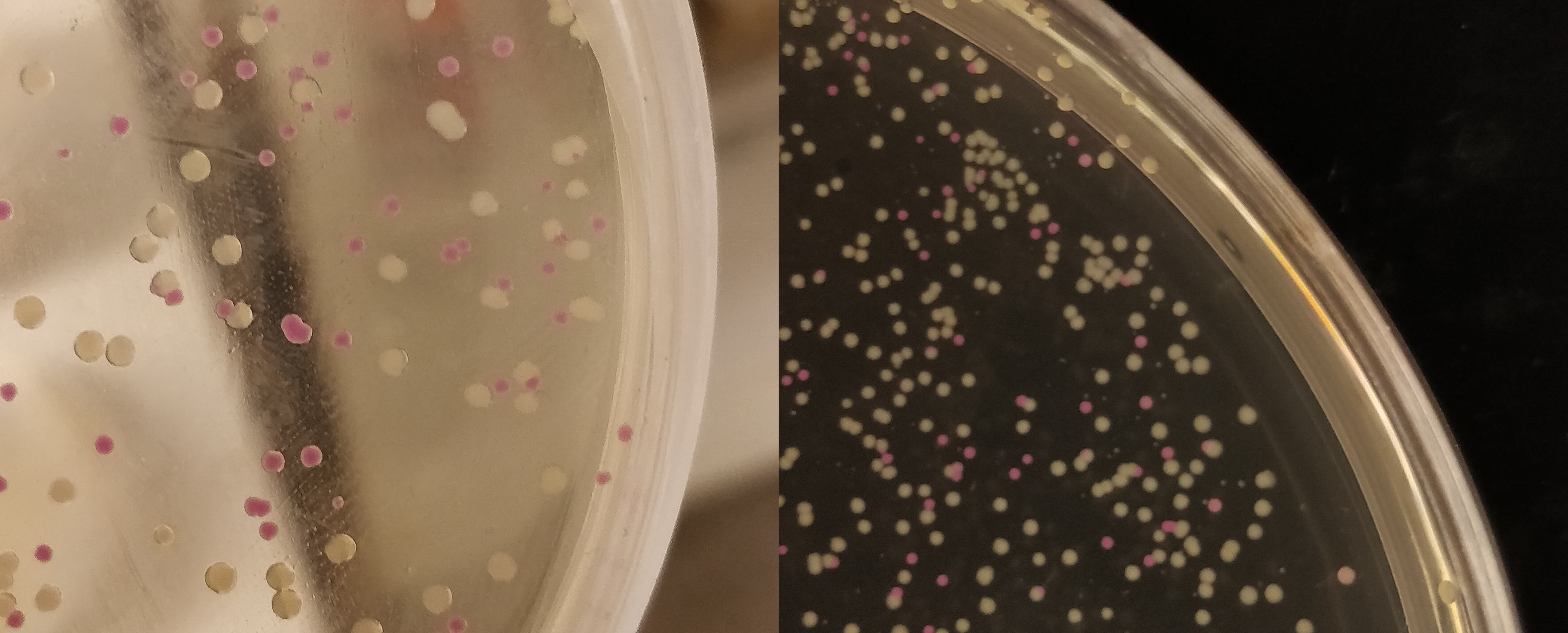
| − | The agar plate contains e.Col (BL21(DE3)) with p.Cons-AsPink (<partinfo>BBa_K3182100</partinfo>) which expresses AsPink and makes the colonies pink. These colonies | + | The agar plate contains e.Col (BL21(DE3)) with p.Cons-AsPink (<partinfo>BBa_K3182100</partinfo>) which expresses AsPink and makes the colonies pink. These colonies were then later used for color screening to see if the ligation was successful. |
[[File:T--Linkoping_Sweden--pinkwhite156.jpeg|700px|thumb|left|<b>Figure 2.</b> E. coli (BL21) cells used for pink-white screening, the cells were incubated for 16 hours in 37 degrees Celsius. <partinfo>BBa_K3182100</partinfo> was cut with BamHI and PstI to remove pCons-AsPink and <partinfo>BBa_K3182006</partinfo> (magainin 2) and <partinfo>BBa_K3182104</partinfo> (CHAP) was ligated into the plasmid. The white colonies indicate a successful ligation. All the colonies that were later colony screened with PCR amplification of the insert and the ampified strand was run on an agarose gel. The gel implied that all screened colonies was successful, i.e. contained <partinfo>BBa_K3182100</partinfo> with magainin 2 / CHAP instead of pCons-AsPink. ]] | [[File:T--Linkoping_Sweden--pinkwhite156.jpeg|700px|thumb|left|<b>Figure 2.</b> E. coli (BL21) cells used for pink-white screening, the cells were incubated for 16 hours in 37 degrees Celsius. <partinfo>BBa_K3182100</partinfo> was cut with BamHI and PstI to remove pCons-AsPink and <partinfo>BBa_K3182006</partinfo> (magainin 2) and <partinfo>BBa_K3182104</partinfo> (CHAP) was ligated into the plasmid. The white colonies indicate a successful ligation. All the colonies that were later colony screened with PCR amplification of the insert and the ampified strand was run on an agarose gel. The gel implied that all screened colonies was successful, i.e. contained <partinfo>BBa_K3182100</partinfo> with magainin 2 / CHAP instead of pCons-AsPink. ]] | ||
| Line 15: | Line 15: | ||
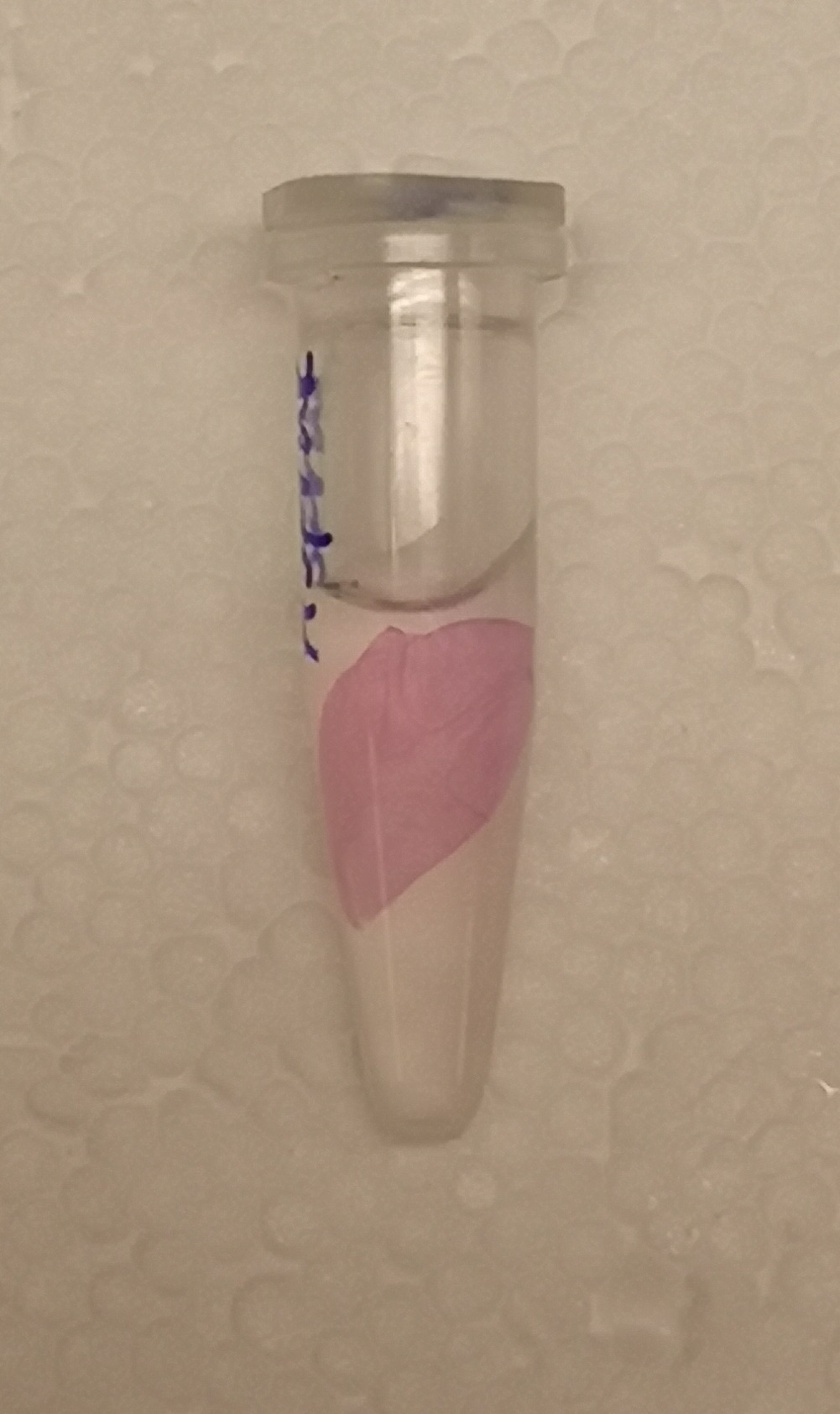
[[File:T--Linkoping Sweden--aspink bunden.jpeg|150px|left|thumb|<b><I>Figure 1.</I></b> Lysate (via sonication) from BL21 E. coli was incubated with Epiprotect (microbial cellulose bandage) for 1h and washed thrice with 70% ethanol.] ]] | [[File:T--Linkoping Sweden--aspink bunden.jpeg|150px|left|thumb|<b><I>Figure 1.</I></b> Lysate (via sonication) from BL21 E. coli was incubated with Epiprotect (microbial cellulose bandage) for 1h and washed thrice with 70% ethanol.] ]] | ||
| − | + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | |
<b>CBD-asPink bindning capacity</b><br> | <b>CBD-asPink bindning capacity</b><br> | ||
Revision as of 09:20, 27 August 2019
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_K1033933
2019 iGEM team Linkoping Sweden
2019 iGEM team Linkoping Sweden validated this part.
Growth of p.cons-asPink
The agar plate contains e.Col (BL21(DE3)) with p.Cons-AsPink (BBa_K3182100) which expresses AsPink and makes the colonies pink. These colonies were then later used for color screening to see if the ligation was successful.
CBD-asPink bindning capacity
To test the bindning capacity of CBD-asPink (BBa_K3182000) the white microbial cellulose bandage was into the sonicated lysate of BL21(DE3) with the expressed fusion protein (see Figure 1) and incubated for 30 minutes.
The bandage was then washed thrice in 70% ethanol which confirmed that the bindning of CDB-asPink was still intact after the washes to the bandage.]
CBD-asPink thrombin cleavage
User Reviews
UNIQcb497a32a138a809-partinfo-00000006-QINU UNIQcb497a32a138a809-partinfo-00000007-QINU