Difference between revisions of "Part:BBa K2728001"
Bluepumpkin (Talk | contribs) (→Experimental Characterization) |
Bluepumpkin (Talk | contribs) (→Improvements) |
||
| Line 105: | Line 105: | ||
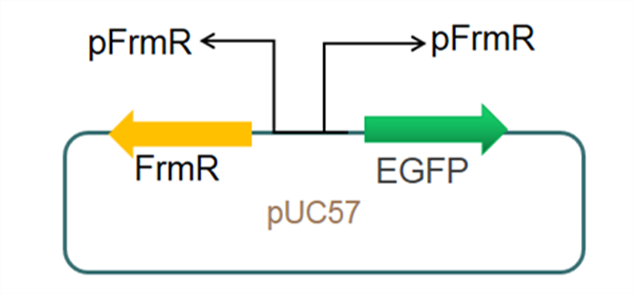
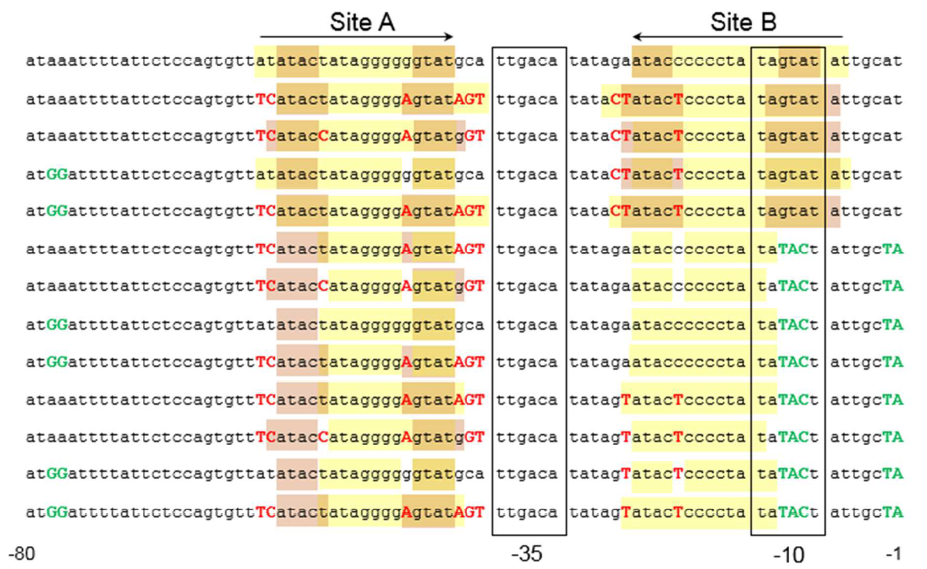
The sequence of this part was taken from the research of Rohlhill J. et al. The 2 binding sites of variations are -35 and -10 (Fig 1). We ordered synthesized plasmid with pFrmR and EGFP from Gensceipt and constructed pFrmR-EGFP-FrmR reporter system on plasmid pUC57. After dosing formaldehyde of 100 to 400uM, we tested the EGFP expression to identify the activity of the formaldehyde induced response of this prompter. | The sequence of this part was taken from the research of Rohlhill J. et al. The 2 binding sites of variations are -35 and -10 (Fig 1). We ordered synthesized plasmid with pFrmR and EGFP from Gensceipt and constructed pFrmR-EGFP-FrmR reporter system on plasmid pUC57. After dosing formaldehyde of 100 to 400uM, we tested the EGFP expression to identify the activity of the formaldehyde induced response of this prompter. | ||
<br /><br /> | <br /><br /> | ||
| − | The research conducted by Rohlhill J. et al. has proved that, pFrmR retains formaldehyde responsiveness, with 2-fold higher GFP expression in response to 100 μM formaldehyde than the native pfrm ([https://parts.igem.org/Part:BBa_K749008 BBa_K749008] )[4].This is quite a new discovery and we have not found other published researches having applied this promoter, which means, we are the first team that brings it to iGEM! | + | The research conducted by Rohlhill J. et al. has proved that, pFrmR retains formaldehyde responsiveness, with 2-fold higher GFP expression in response to 100 μM formaldehyde than the native pfrm ([https://parts.igem.org/Part:BBa_K749008 BBa_K749008], submitted by TMU-Tokyo in 2012 and they did not observe any GFP expression that year.)[4].This is quite a new discovery and we have not found other published researches having applied this promoter, which means, we are the first team that brings it to iGEM! |
<br /> | <br /> | ||
<br /> | <br /> | ||
Revision as of 00:51, 18 October 2018
pfrmR - An Engineered Formaldehyde-Inducible Promoter
Basic Description
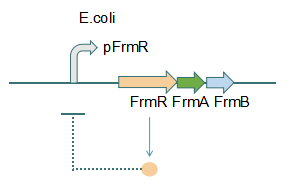
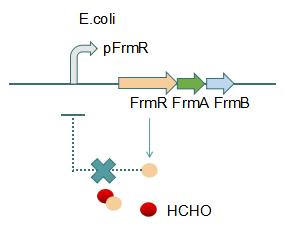
This promoter is an engineered formaldehyde-inducible promoter. Escherichia coli has a native formaldehyde-inducible promoter, pfrm, which is found upstream of the frmRAB formaldehyde detoxification operon. FrmR, the first product of the operon, is a member of the DUF156 family of DNA-binding transcriptional regulators. It binds the frmRAB promoter region and is negatively allosterically modulated by formaldehyde. FrmR is specific to formaldehyde, responding to acetaldehyde, methylglyoxal, and glyoxal to far lesser degrees and not at all to a range of other aldehydes and alcohols tested. The genes frmA and frmB encode a formaldehyde dehydrogenase and S-formylglutathione hydrolase, respectively, and are responsible for detoxifying formaldehyde to formic acid in a glutathione-dependent pathway. The negative-feedback regulation of the frmRAB operon is similar to that of many other prokaryotic operons, whereby the transcription factor represses its own transcription.
Fig 1: Without Formaldehyde
Fig 2: With Formaldehyde
Features
- It’s a formaldehyde-inducible promoter from E.coli.
- It’s an engineered promoter. It retains formaldehyde responsiveness, with 2-fold higher GFP expression in response to 100 μM formaldehyde than the native pfrm. Application of this promoter with higher basal and induced expression levels before methanol assimilation genes achieves higher biomass titers than the native E. coli pfrm.
Origins
Escherichia coli
Experimental Characterization
Though we failed a lot of times that we cannot even count, due to a lot of different reasons and controls. Eventually we found the proper setting of our equipment, and got some promising data as below:
Experiment 1
Culture environment: saturated formaldehyde aqueous solution with concentration of 37 percent
Instruments: tecan infinite m1000
Experiment group: BL21 with pFrmR+EGFP
Control group: BL21 (with no plasmid transformed)
- Test the OD number of overnight-cultured strain;
- Diluted to 0.05, the strains are cultured under 37 celsius, 220prm.
- Take 5 tubes of 1ml system both from experiment group and control group and add 0ul, 1ul, 2.5ul, 5ul, and 7.5ul saturated formaldehyde solution into each tube. Mix evenly.
- ake 150ul from each of the tubes into 96 orifice plate.
- est fluorescence in tecan infinite m1000.
Related conditions:
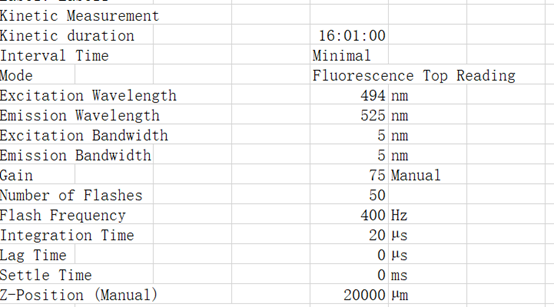
Excitation Wavelength 495 nm
Emission Wavelength 525 nm
List of actions in this measurement script:
Shaking (Orbital) Duration: 900 s
Shaking (Orbital) Amplitude: 2 mm
Shaking (Orbital) Frequency: 306 rpm
Results:
In solution with 1ul of formaldehyde, luminescence was the most obvious, indicating more active pFrmR.
Experiment 2
Culture environment: saturated formaldehyde aqueous solution with concentration of 37 percent
Instruments: tecan infinite m1000
Experiment group: BL21 with pFrmR+EGFP
Control group: BL21
- Test the OD number of overnight-cultured strain;
- Diluted to 0.05, the strains are cultured under 37 celsius, 220prm.
For experiment group:
- Take 5 tubes of 1ml system both from experiment group and control group and add 0ul,0.2ul,0.5ul,1ul,and 1.5ul saturated formaldehyde solution into each tube. Mix evenly.
- Take 150ul from each of the tubes into 96 orifice plate.
- Test fluorescence in tecan infinite m1000.
Related conditions:
Excitation Wavelength 495 nm
Emission Wavelength 525 nm
List of actions in this measurement script:
Shaking (Orbital) Duration: 900 s
Shaking (Orbital) Amplitude: 2 mm
Shaking (Orbital) Frequency: 306 rpm
Results:
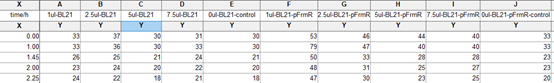
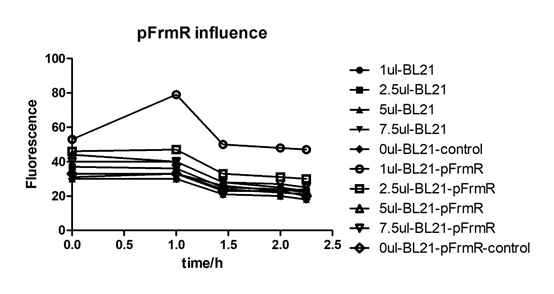
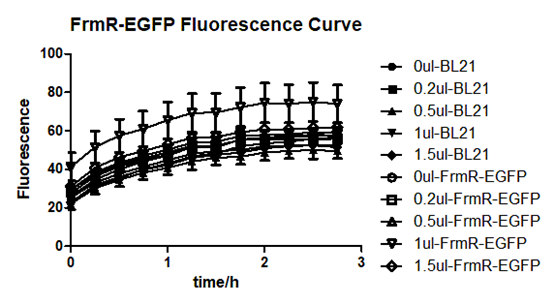
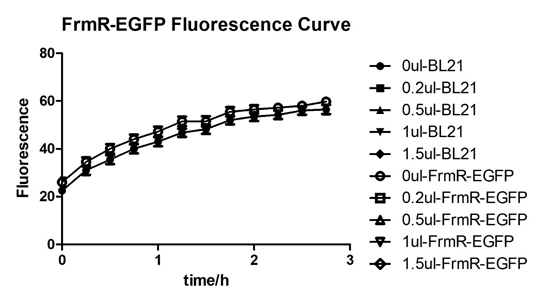
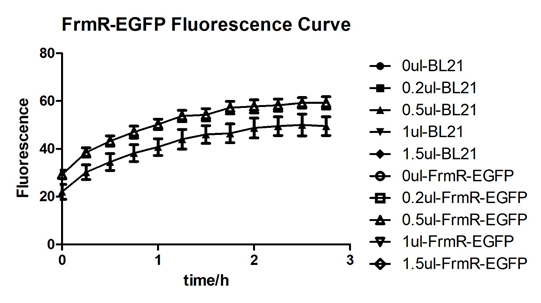
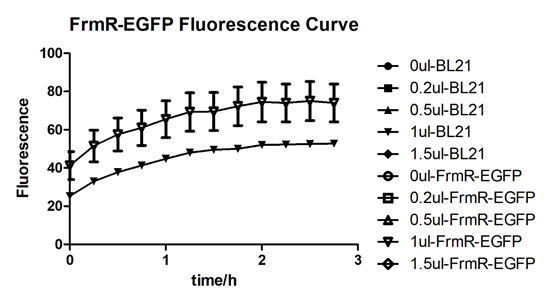
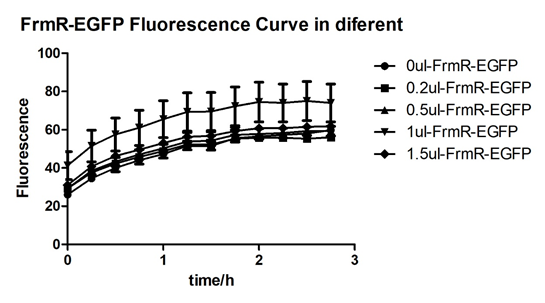
Summarized graph for all groups of FrmR and BL21. In solution with 1ul of formaldehyde, luminescence was the most obvious, indicating more active pFrmR.
In solution with no formaldehyde dosed, BL21 with EGFP had similar expression of fluorescence with control group.
In solution with addition of 0.2ul of formaldehyde, BL21 with EGFP had similar expression of fluorescence with the control group, which means in this condition the activity of pFrmR was low.
In solution with addition of 0.5ul of formaldehyde, the difference in fluorescent degree between BL21 with EGFP and the control group was clear, indicating higher activity of pFrmR.
In solution with addition of 1ul of formaldehyde, the difference in fluorescent degree between BL21 with EGFP and the control group was the most obvious, indicating highest activity of pFrmR which successfully activated EGFP.
In solution with addition of 1.5ul formaldehyde solution, the difference in fluorescent degree between BL21 with EGFP and the control group was clear, indicating the certain level of activity of pFrmR.
In solution with addition of 1ul of formaldehyde solution, the activity of pFrmR was the highest.
In formaldehyde free condition, FrmR inhibiting factor regulates pFrmR through negative feedback, lowering the activity of the promotor. After adding the inductor, FrmR inhibiting factor combined with formaldehyde molecules, lowering the effect of the negative feedback, thus the activity of pFrmR will be higher.
Improvements
The sequence of this part was taken from the research of Rohlhill J. et al. The 2 binding sites of variations are -35 and -10 (Fig 1). We ordered synthesized plasmid with pFrmR and EGFP from Gensceipt and constructed pFrmR-EGFP-FrmR reporter system on plasmid pUC57. After dosing formaldehyde of 100 to 400uM, we tested the EGFP expression to identify the activity of the formaldehyde induced response of this prompter.
The research conducted by Rohlhill J. et al. has proved that, pFrmR retains formaldehyde responsiveness, with 2-fold higher GFP expression in response to 100 μM formaldehyde than the native pfrm (BBa_K749008, submitted by TMU-Tokyo in 2012 and they did not observe any GFP expression that year.)[4].This is quite a new discovery and we have not found other published researches having applied this promoter, which means, we are the first team that brings it to iGEM!
Future Improvements:
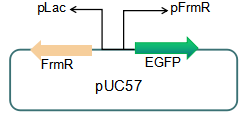
We plan to optimize our reporter vector by introducing an independent promoter pLac to regulate the expression of FrmR.
Fig 1: Sites of mutants
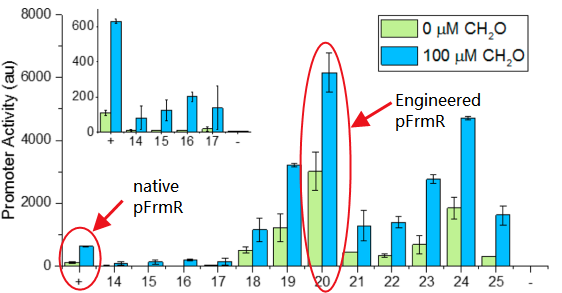
Fig 2: Comparison of activity
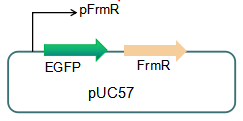
Fig 3: Current reporter system
Fig 5: Future reporter system
Potential Application
- To construct a formaldehyde sensor with this promoter.
- To enable higher growth under formaldehyde pressure with the application of the engineered formaldehyde responsive promoter.
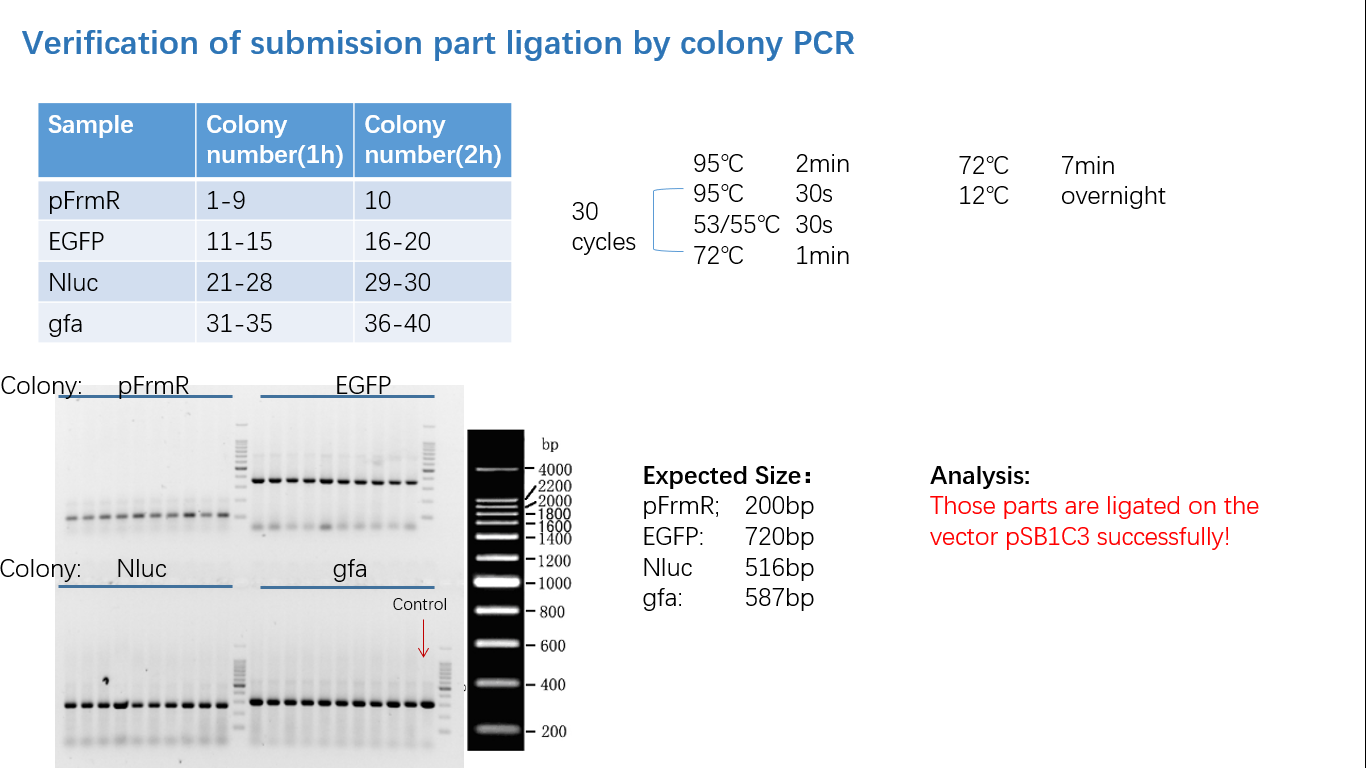
Parts Verification Before Submission
We verified our parts in the lab before submission. They are reliable! Please feel free to apply them onto your project.=)
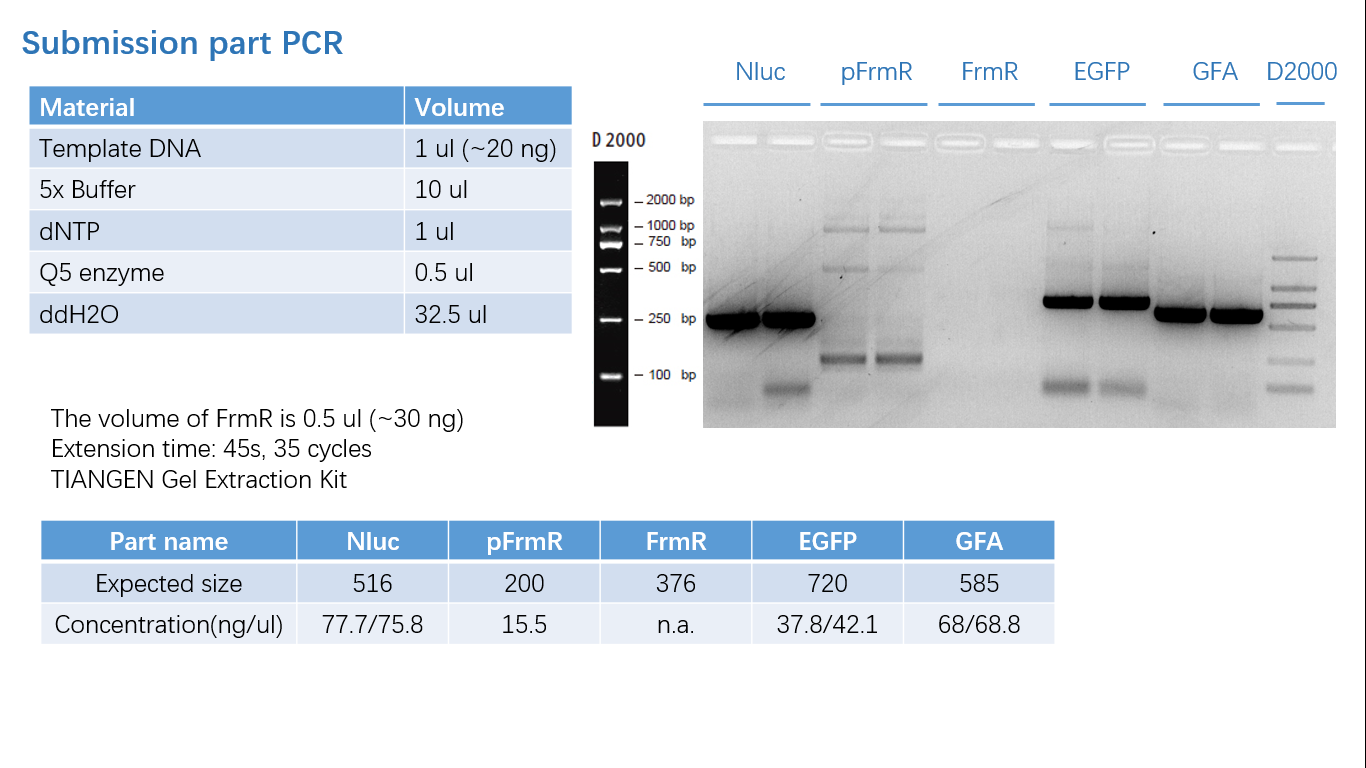
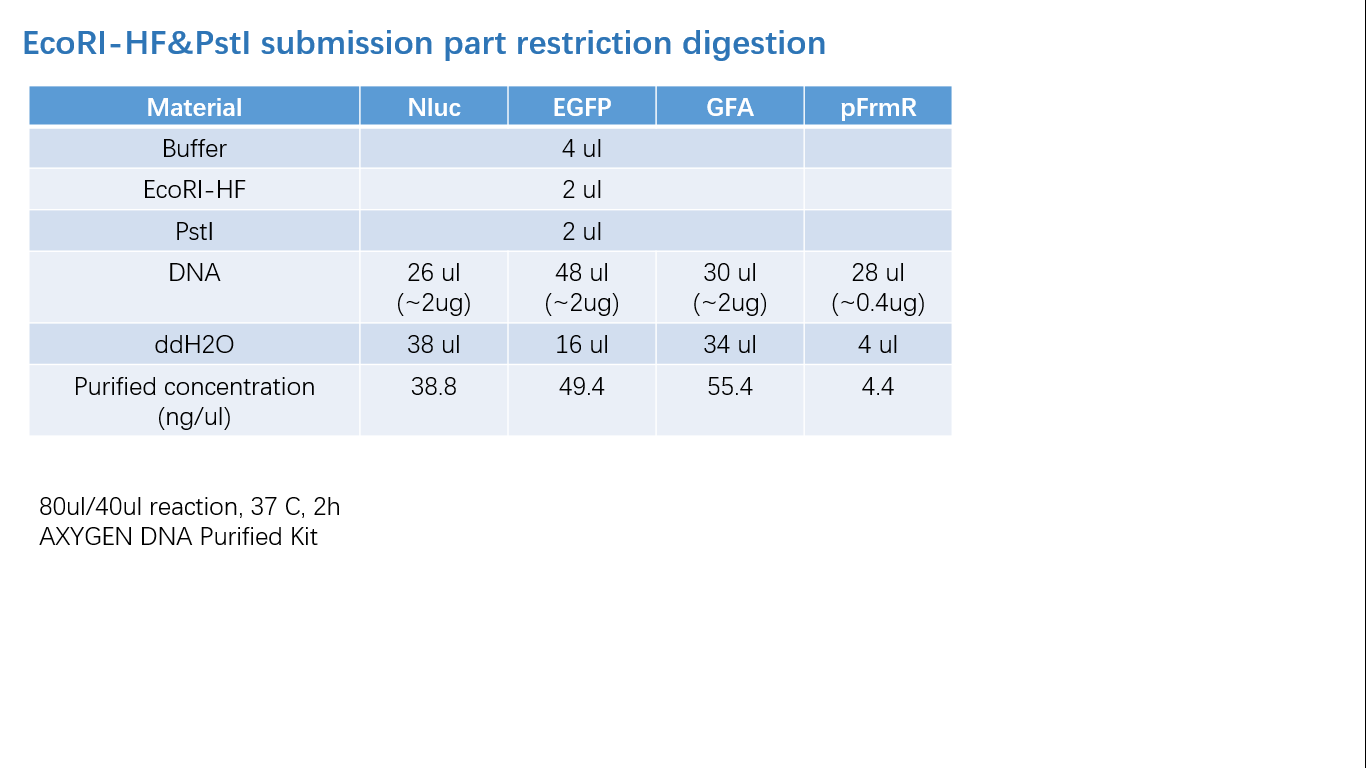
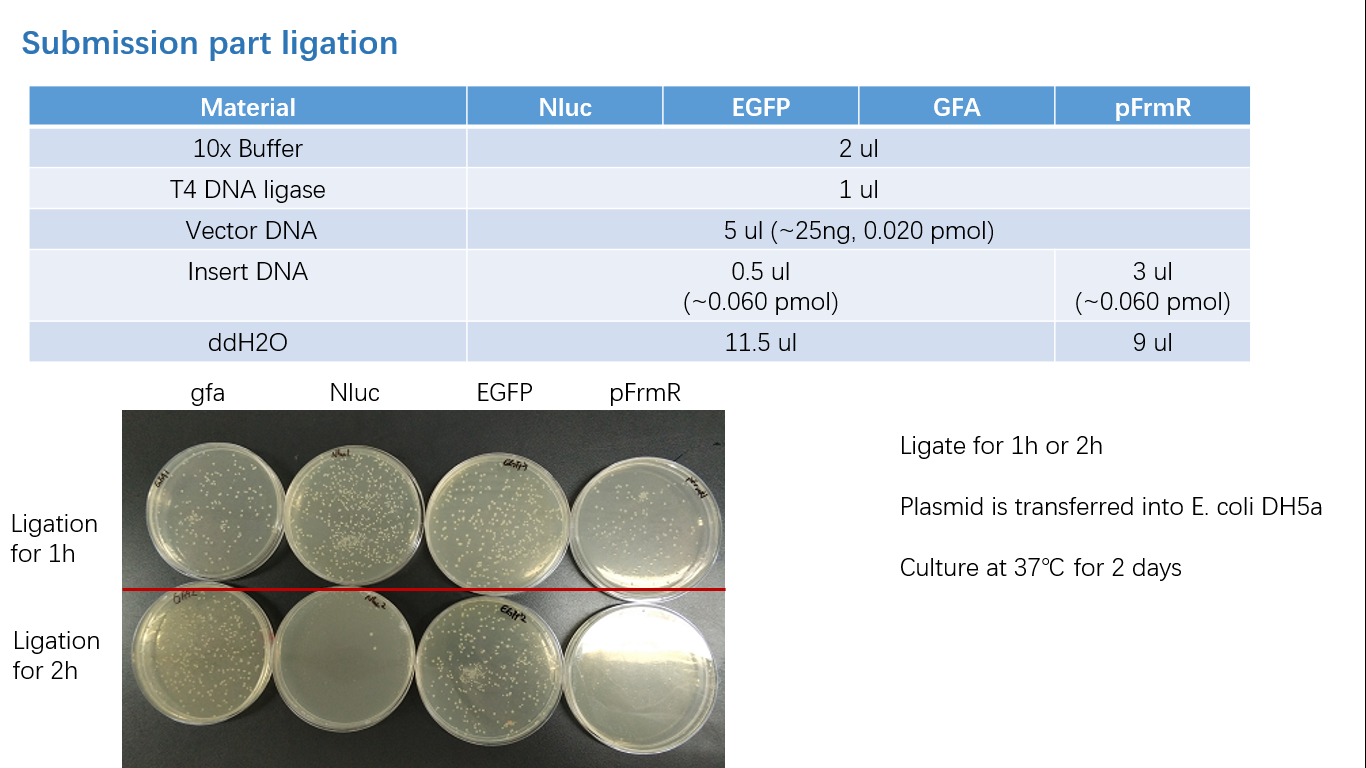
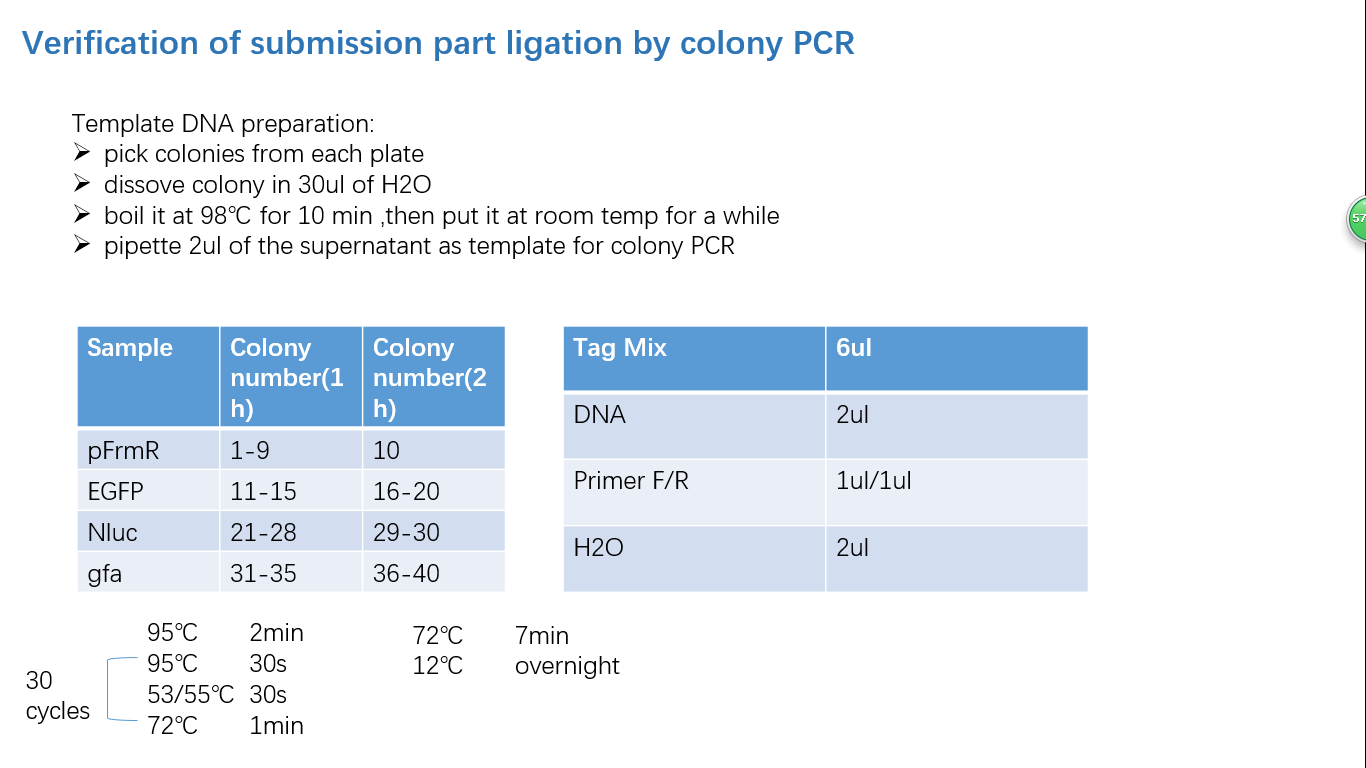
Fig 1: PCR (to get targeted genes)
Fig 2: Restriction Digestion
Fig 3: Ligation
Fig 4: Colony PCR
Fig 5: Gel Verification
References
- Osman, D., Piergentili, C., Chen, J., Sayer, L. N., Uson, I., Huggins, T. G., Robinson, N. J., and Pohl, E. (2016) The Effectors and Sensory Sites of Formaldehyde-Responsive Regulator FrmR and Metal-Sensing Variant. J. Biol. Chem. 291, 19502-19516
- Denby, K. J., Iwig, J., Bisson, C., Westwood, J., Rolfe, M. D., Sedelnikova, S. E., Higgins, K., Maroney, M. J., Baker, P. J., Chivers, P. T., and Green, J. (2016) The mechanism of a formaldehyde-sensing transcriptional regulator. Sci. Rep. 6, 38879
- Gonzalez, C. F., Proudfoot, M., Brown, G., Korniyenko, Y., Mori, H., Savchenko, A. V., and Yakunin, A. F. (2006) Molecular basis of formaldehyde detoxification: Characterization of two S-formylglutathione hydrolases from Escherichia coli, FrmB and YeiG. J. Biol. Chem. 281, 14514-14522
- Rohlhill J, Sandoval N R, Papoutsakis E T. Sort-seq approach to engineering a formaldehyde-inducible promoter for dynamically regulated Escherichia coli growth on methanol.[J]. Acs Synthetic Biology, 2017, 6(8)
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]