Difference between revisions of "Part:BBa K2886010"
| Line 11: | Line 11: | ||
[[File:T--Fudan-CHINA--figuer1for010.jpg|600px|thumb|left|'''Figure 1:''' The structure and ik_ball _wtd of wild type and mutant type tet binding site. (a) Structure of wild type (WT) tet binding site. (b) Structure of mutant type (MT) tet binding site. The transcription factor is colored to show its electrostatics distribution. Compared to WT, MT with a substitution of A to C on base pair 11 has a lower interface energy decreasing from-37.467 to -38.411 kcal/mol, tested by RosettaDock. (c) The most significant improvement is lk_ball_wtd (also known as orientation-dependent solvation energy), indicating MTmay have a higher affinity to the transcriptional factor.]] | [[File:T--Fudan-CHINA--figuer1for010.jpg|600px|thumb|left|'''Figure 1:''' The structure and ik_ball _wtd of wild type and mutant type tet binding site. (a) Structure of wild type (WT) tet binding site. (b) Structure of mutant type (MT) tet binding site. The transcription factor is colored to show its electrostatics distribution. Compared to WT, MT with a substitution of A to C on base pair 11 has a lower interface energy decreasing from-37.467 to -38.411 kcal/mol, tested by RosettaDock. (c) The most significant improvement is lk_ball_wtd (also known as orientation-dependent solvation energy), indicating MTmay have a higher affinity to the transcriptional factor.]] | ||
<br style="clear: both" /> | <br style="clear: both" /> | ||
| + | |||
| + | For more detailed information, please click here[[http://2018.igem.org/Team:Fudan-CHINA/Improve]] to read our wiki. | ||
===Improved from a previous part=== | ===Improved from a previous part=== | ||
Revision as of 20:32, 17 October 2018
tet binding site
TetR is a kind of repressible promoter commonly used in synthetic biology. This time, we designed a mutation of TetR form Part:BBa_R0040 named pTight and according to the simulative result, the pTight shows a higher bind affinity compared to the original TetR.
Usage and Biology
BBa_K2886010 is a repressible tet binding site improved from BBa_R0040, a commonly used part in iGEM. This time, we designed a mutation of tet binding site. According to the simulative result, BBa_K2886010 shows a higher bind affinity compared to the original TetR. Figure 6 exhibits the result.
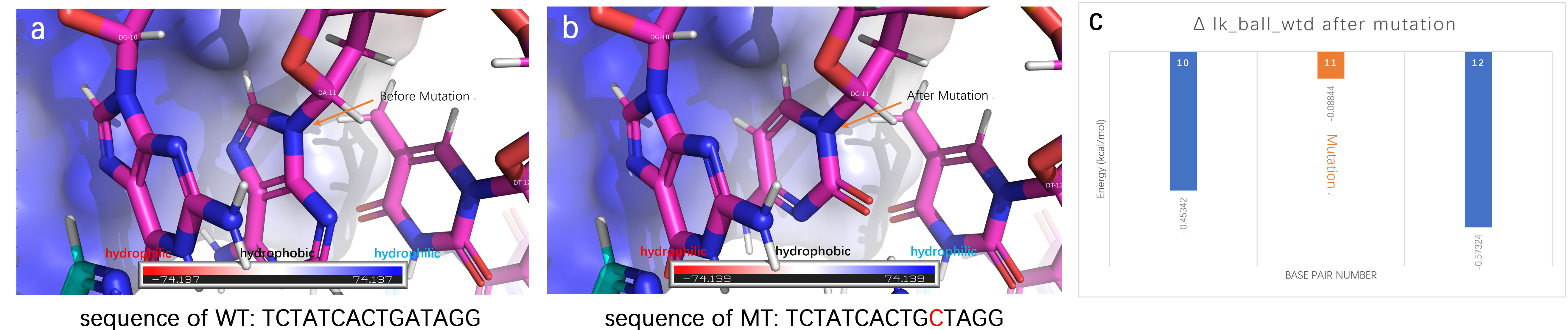
For more detailed information, please click herehttp://2018.igem.org/Team:Fudan-CHINA/Improve to read our wiki.
Improved from a previous part
Part:BBa_R0040 is a commonly used part in iGEM which has over 1000 uses till 2017. And this year, Fudan-CHINA makes a mutation to Part:BBa_R0040 to improve its combination with the tTA transcriptional factor.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
