Difference between revisions of "Part:BBa K2705006"
| Line 41: | Line 41: | ||
==Proof of Function== | ==Proof of Function== | ||
| − | Vector pHT01-P | + | Vector pHT01-P<sub><i>gltAB</i></sub>-LacI-P<sub><i>grac</i></sub>-TetA was converted into LL3, and correct transformants were fermented in M9 culture medium with different extracellular glutamate concentrations (0, 2.5 and 5g/L). From the 6th hour, we tested bacteria with several assays every 3 hours. LL3 Δ<i>bam</i> strain was chosen as control system. |
===Expression level of <i>tetA</i> by microplate assay=== | ===Expression level of <i>tetA</i> by microplate assay=== | ||
Revision as of 13:41, 17 October 2018
PgltAB-LacI-Pgrac-TetA
LacI is an inhibitor of Pgrac, TetA is a tetracycline resistance protein. The LacI expression is downregulated by glutamate because PgltAB is upregulated by GltC, which is repressed by cellular glutamate level; Pgrac is down regulated by LacI. As a result, higher the cellular gultamate level is, more TetA expressed, better the bacteria can survive under tetracycline circumstance. A BamHI cutting site and a 5bp non-coding site were left in the part during construction of the vector, they had no influence in the expected function.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 1229
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Introduction
Bacillus amyloliquefaciens LL3 and poly-γ-glutamate(γ-PGA)
B. amyloliquefaciens LL3 is one of most prevalent Gram-positive aerobic spore-forming bacteria. It was isolated from fermented food with the ability to produce poly-γ-glutamate(γ-PGA) in a glutamate-independent way.
γ-PGA is a commercially available biopolymer made of D- or L-glutamate units connected by amide linkages, which is nontoxic, edible, degradable and absorbent. It can be used as food and cosmetic additives as well as flocculants for sewage disposal.
Here, we chose B. amyloliquefaciens LL3 as our genetically engineered microorganism (GEM) and γ-PGA as the target product. We intended to enhance the yield of γ-PGA in LL3 with our PopQC system. Besides, the PopQC system also has great potential to be utilized in other bacterial strains.
Principle of Population Quality Control (PopQC) system
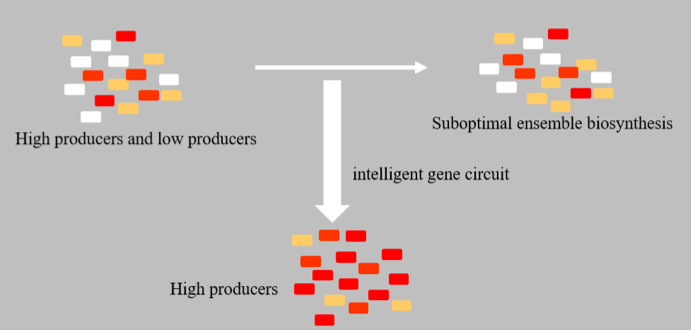
Population Quality Control (PopQC) system is a new approach designed for biosynthesis yield enhancement, based on non-genetic cell-to-cell variation, which includes unequal cell division, different gene copy numbers, epigenetic modifications, random gene expression, volatile RNA stability, protein activity, etc. Because of those differences, different cells in a single colony will have considerable variations in protein and metabolite concentrations. Therefore, in cell cultures there will be both high- and low-producers, and the intrinsic low-producers might cause suboptimal ensemble biosynthesis.
The elimination of low-producers can realize the efficient utilization of substrates and high yield of target products. Thus, it has been proved to be an efficient way for biosynthesis being more suitable for large-scale industrial production. In our project, PopQC was designed as a plasmid-based gene circuit in B. amyloliquefaciens LL3, which continuously selects high-performing cells in order to further improve the yield of target metabolite—glutamate, and then the secondary metabolites—γ-PGA. (See Figure 1.)
In our work, promoter PgltAB(BBa_K2705000), promoter Pgrac(BBa_K2705002), lacI(BBa_K2705001) gene and tetA(BBa_K2705003) gene were composed to build up the system. (See Figure 2.) The function of PgltAB is documented in BBa_K2705000.
In Bacillus amyloliquefaciens LL3 exists the glt operon, which is responsible for intracellular glutamate synthesis (See BBa_K2705000 for more details about PgltAB). TetA is a tetracycline resistance protein[TetA(C) inner-membrane-associated protein] (See BBa_K2705007 and BBa_K2705003 for more details about TetA).
With a specific extracellular tetracycline concentration, when intracellular glutamate-precursor of γ-PGA-concentration of the individual is low, GltC level will go up, which activates the PgltAB to express lacI. LacI furthermore represses Pgrac and as a result, represses tetA expression. On the contrary, for high-producers, the concentration of intracellular GltC will go down, which represses the PgltAB to express lacI, and the tetA expression is not affected. Therefore, high-producers will synthesize enough amount of tetracycline efflux pumps to maintain alive while low-producers won't be able to survive. Consequently, the average intracellular glutamate concentration among the population is enhanced, which will finally lead to γ-PGA yield enhancement in LL3.
Proof of Function
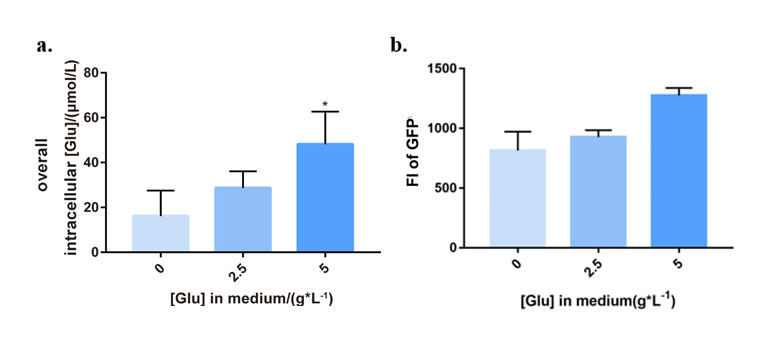
Vector pHT01-PgltAB-LacI-Pgrac-TetA was converted into LL3, and correct transformants were fermented in M9 culture medium with different extracellular glutamate concentrations (0, 2.5 and 5g/L). From the 6th hour, we tested bacteria with several assays every 3 hours. LL3 Δbam strain was chosen as control system.
Expression level of tetA by microplate assay
To test the expression of tetA, we tagged it with the fluorescent reporter GFP-coding gene (BBa_K2705004), whose expression was detected by microplate assay (395nm\509nm). The intracellular glutamate concentration and bacteria concentration (OD600) were also examined, respectively. (See Figure 3.) It could be concluded that with the increasing glutamate in medium, intracellular glutamate concentration went high, and tetA of PopQC was upregulated to express. The results suggested that the system can help individuals with higher intracellular glutamate concentration express more TetA, so that be able to survive in the tetracycline condition.
Measurement of γ-PGA yield
After 24 hours fermentation, the γ-PGA yields of both strains without extra glutamate were tested. With PopQC system, NK-Ipop strain produced more γ-PGA, which approved the system function. See Figure 4.

References
Weitao G, Mingfeng C, Cunjiang S et al. Complete Genome Sequence of Bacillus amyloliquefaciens LL3, Which Exhibits Glutamic Acid-Independent Production of Poly-γ-Glutamic Acid. J Bacteriol. 2011 Jul; 193(13): 3393–3394.
Picossi S, Belitsky B R, Sonenshein A L. Molecular mechanism of the regulation of Bacillus subtilis gltAB expression by GltC[J]. J Mol Biol., 2007, 365(5):1298-1313.
Commichau FM, Herzberg C, Tripal P et al. A regulatory protein-protein interaction governs glutamate biosynthesis in Bacillus subtilis: the glutamate dehydrogenase RocG moonlights in controlling the transcription factor GltC. Mol Microbiol. 2007 Aug;65(3):642-654.
Bohannon D E and Sonenshein A L. Positive regulation of glutamate biosynthesis in Bacillus subtilis. J Bacteriol. 1989 Sep; 171(9): 4718–4727.
Xiao Y, Bowen CH, Liu D, Zhang F. Exploiting nongenetic cell-to-cell variation for enhanced biosynthesis. Nat Chem Biol. 2016 May;12(5):339-344.
Brodersen DE1, Clemons WM Jr, Carter AP, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell. 2000 Dec 22;103(7):1143-1154.