Difference between revisions of "Part:BBa K2675042"
m |
|||
| Line 64: | Line 64: | ||
| − | + | ===References=== | |
| + | [1] Erez Z, Steinberger-Levy I, Shamir M, Doron S, Stokar-Avihail A, Peleg Y, Melamed S, Leavitt A, Savidor A, Albeck S, Amitai G, Sorek R. Communication between viruses guides lysis-lysogeny decisions. Nature (2017) 541, 488-493. | ||
| + | [2] Espah Borujeni A, Channarasappa AS, Salis HM. Translation rate is controlled by coupled trade-offs between site accessibility, selective RNA unfolding and sliding at upstream standby sites. Nucleic Acids Res (2014) 42, 2646-2659. | ||
| − | + | [3] Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotechnol (2009) 27, 946-50. | |
| − | + | ||
| − | + | [4] Hall BG. Activation of the bgl operon by adaptive mutation. Mol Biol Evol (1998) 15, 1-5. | |
| − | + | ||
| − | + | ||
<!-- --> | <!-- --> | ||
Revision as of 13:34, 17 October 2018
OmpA-SAIRGA expression cassette
This part is the mature arbitrium peptide of phage phi3T (SAIRGA) preceded by the OmpA secretion signal (BBa_K2675002), equipped with a synthetic RBS (BBa_K2675012) and placed under the control of the constitutive strong promoter BBa_J23100 and of the strong terminator L3S2P21 (BBa_K2675031).
This part is functional in E. coli and behaves as predicted: it is able to produce and release the mature hexapeptide SAIRGA in the culturing media of E. coli cells. Thus, it is an improvement of BBa_K2279001 that is not functional in E. coli under an equivalent transcriptional and translational control (BBa_K2675041) (see figure 7 bellow).
Usage and Biology
The hexapeptide SAIRGA is the signalling molecule used for cell-to-cell communication [1]. To perform its quorum sensing function in the natural phi3T phage infection, SAIRGA needs to be secreted out of the cell. It is produced as an immature pre-pro-peptide, AimP (BBa_K2279001 and BBa_K2675001), that upon secretion is cleaved extracellularly to remove the secretion signal and release the mature hexapeptide. The mature peptide then enters cells and binds to its receptor protein AimR (BBa_K2279000 and BBa_K2675000). Binding of SAIRGA to AimR blocks the activator function of AimR that, in turn, facilitates a switch from lytic-to-lysogenic viral cycle. By acting as a negative regulator of AimR, the SAIRGA signal makes the lysis-to-lysogeny switch of phi3T phage dependent on the “quorum” of phi3T phage in Bacillus cells.
In phi3T phage, SAIRGA is produced with specific secretion and protease-cleavage tags [1] that allow secretion and extracellular processing by Bacillus. Since we had no further information about this protease, we could not investigate the ability of Escherichia coli to produce a similar enzyme. Consequently, to produce SAIRGA in E. coli, we have decided to replace this tag by the OmpA secretion signal know to function in E. coli.
To express the OmpA-SAIRGA in E. coli, the sequence was codon optimized for E. coli DH5α, a specific RBS (BBa_K2675012) was designed with the Salis Lab RBS Calculator [2, 3] and the peptide was placed under the control of the constitutive strong promoter (BBa_J23100) and of the strong terminator L3S2P21 (BBa_K2675031). Thus, this composite part (BBa_K2675042) was generated.
This SAIRGA expression part behaved as predicted: it was able to produce and release the mature hexapeptide SAIRGA in the culturing media of E. coli cells.
Experimental validation
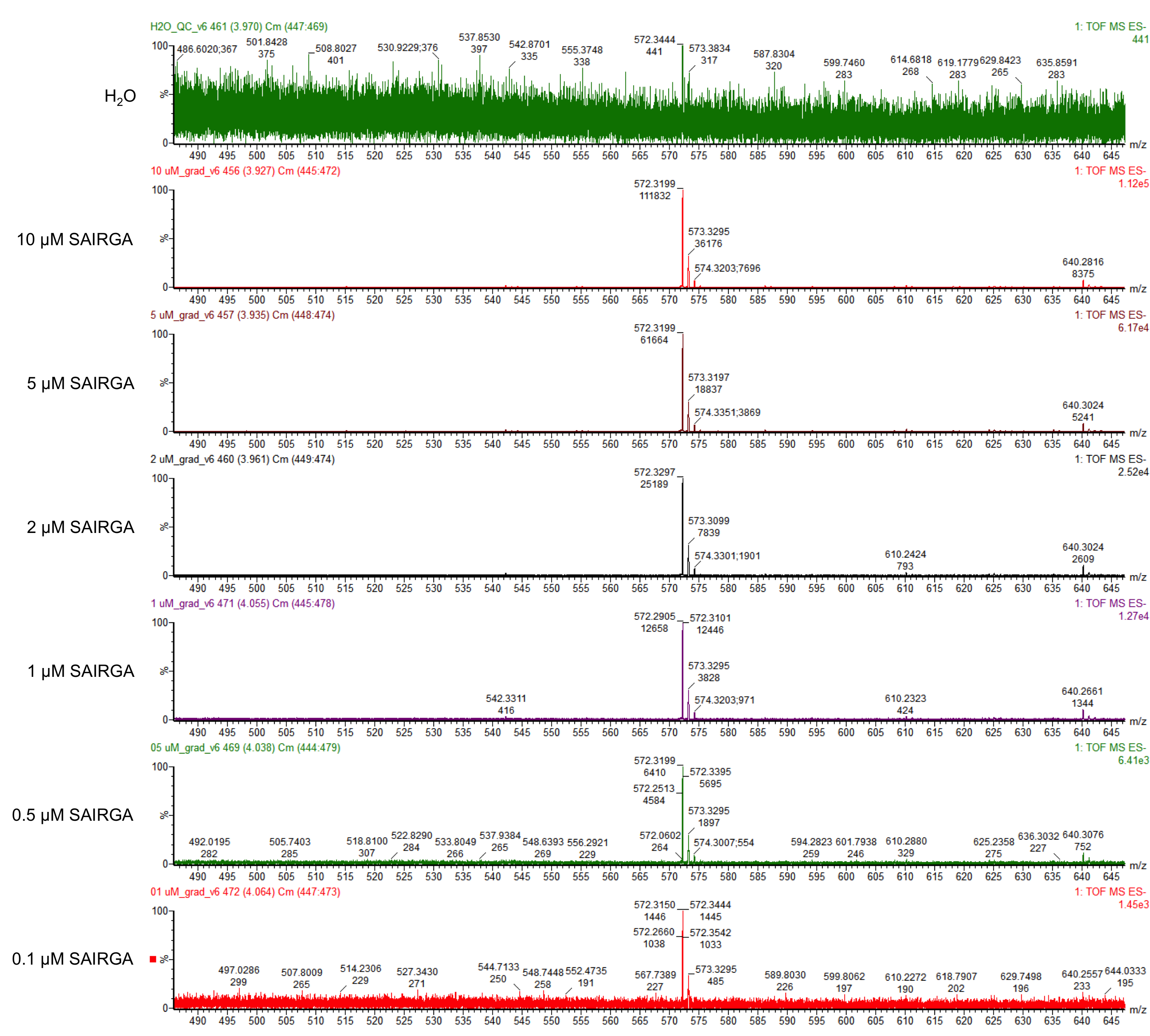
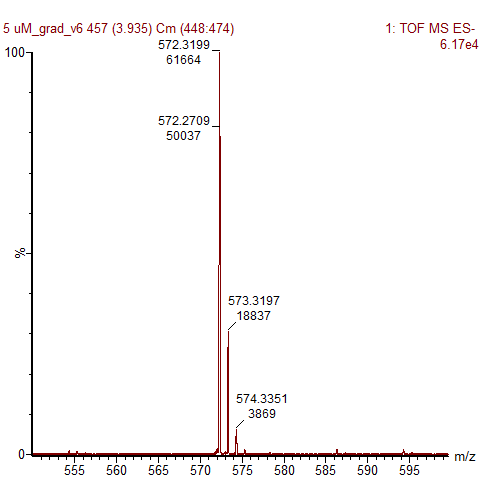
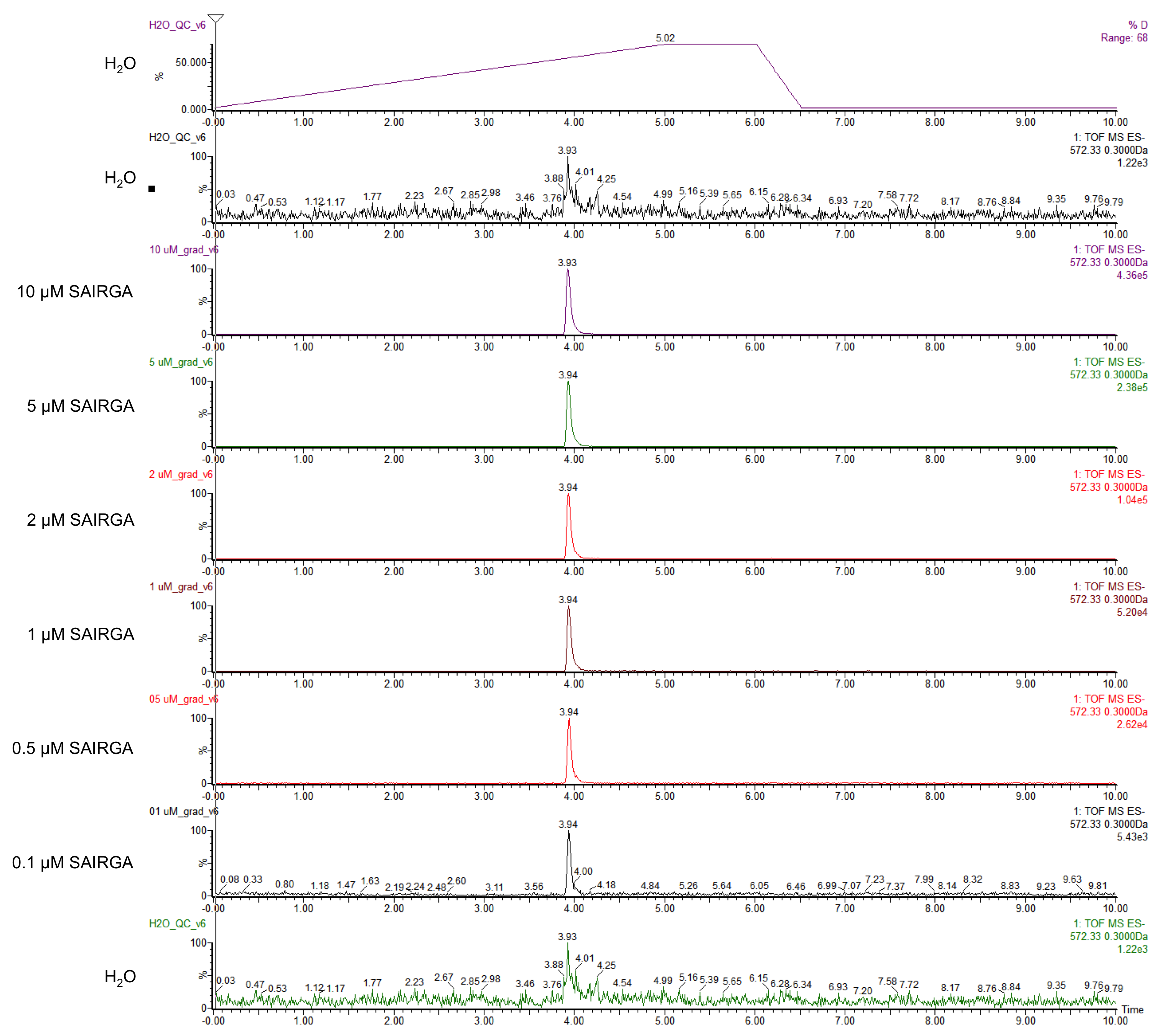
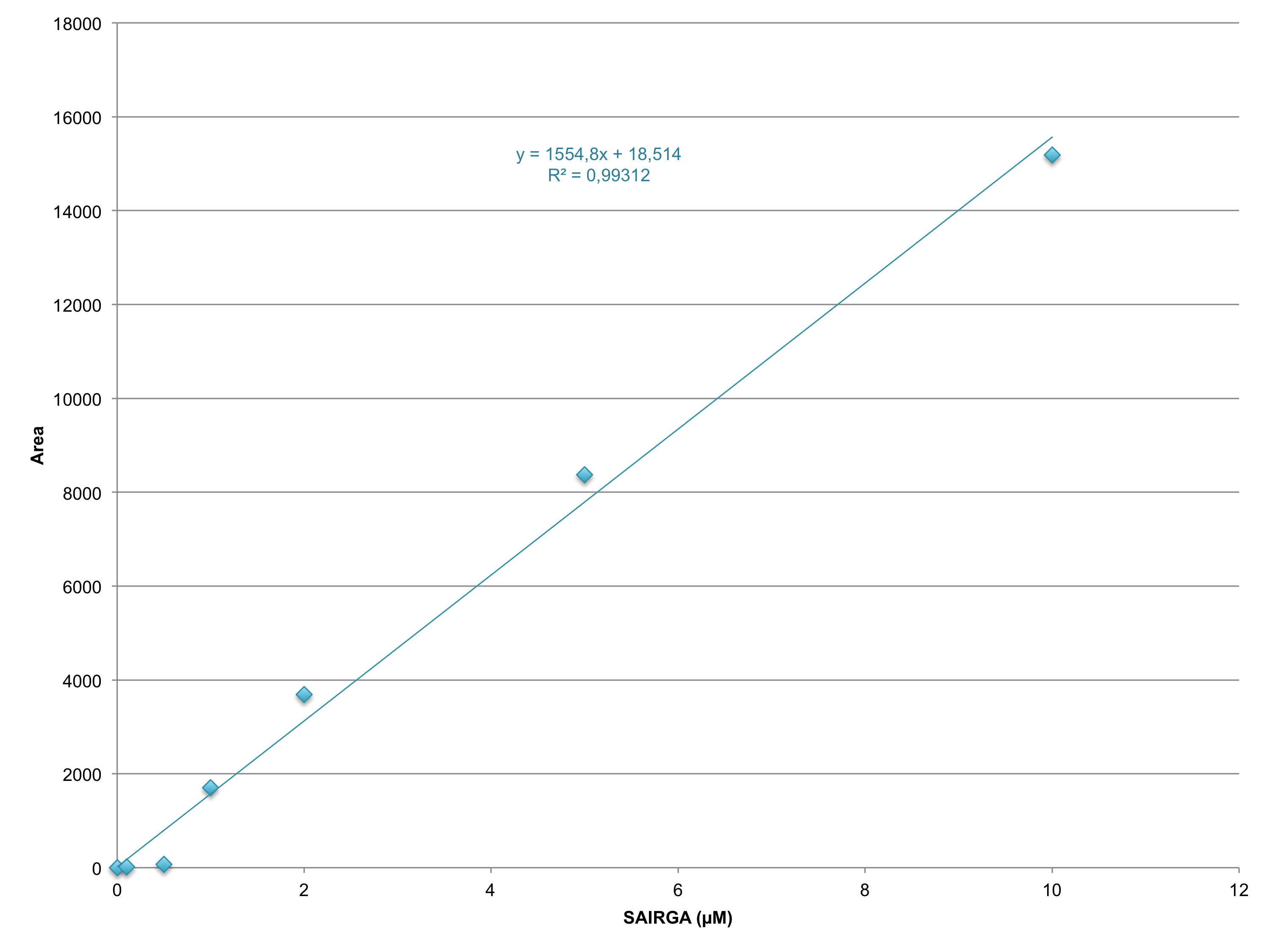
E. coli DH5α harboring this SAIRGA expression plasmid were grown at 30°C and 37°C in mineral salts medium [4] containing 2 mg/mL glucose and 35 μg/mL chloramphenicol. During the culturing, samples were prelevated, centrifuged and the supernatant was first sterilised by filtering through a 0.22 µm membrane, then loaded on a Pall Nanosep centrifugal device with Omega membrane (MWCO 3 kDa). The thus recovered low molecular weight fraction of the culturing media was analysed by LC-MS: ultra high pressure liquid column chromatography (UPLC) on a ACQUITY HSS T3 1.8 µM, 2.1 x 100 mm column (Waters) coupled with a Xevo G2-S Qtof mass spectrometer (Waters). The chromatography was performed using a gradient of using a (A) 1% formic acid in whater / (B) 1% formic acid in acetonitrile with the following program: from 0 to 5 min solvent B was increased from 2% to 70%, then it was maintained for 1 min at 70%; aftreward it was decreased in 30 sec to 2% and it was maintained at 2% for 3.5 min for column re-equilibration. The flow rate was 0.2 mL/min and the column temperature was set to 45°C. The ions were detected in resolution negative mode. The pure synthetic SAIRGA peptide was readily detected by LC-MS. As shown in figures 1 and 2, a peak of 572 Da, which correspond to SAIRGA’s monoisotopic mass [M-H]- was observed at a retention time of 3.9 min (figure 3). The area under the curve is proportional to the concentration of SAIRGA (figure 4).
Figure 1: Mass spectrometry analysis of SAIRGA’s fraction obtained with various amounts of pure synthetic SAIRGA peptide.
Figure 2: Mass spectrometry analysis of SAIRGA’s fraction obtained with 5 µM of pure synthetic SAIRGA peptide (zoom on SAIRGA’s peak).
Figure 3: UPLC chromatograms obtained with various amount of pure synthetic SAIRGA peptide.
Figure 4: Standard curve of the area under the curve obtained by UPLC as a function of the concentration of SAIRGA.
LC-MS analysis of samples isolated from the fermentation broth of the E. coli strains harbouring this SAIRGA expression part was performed.
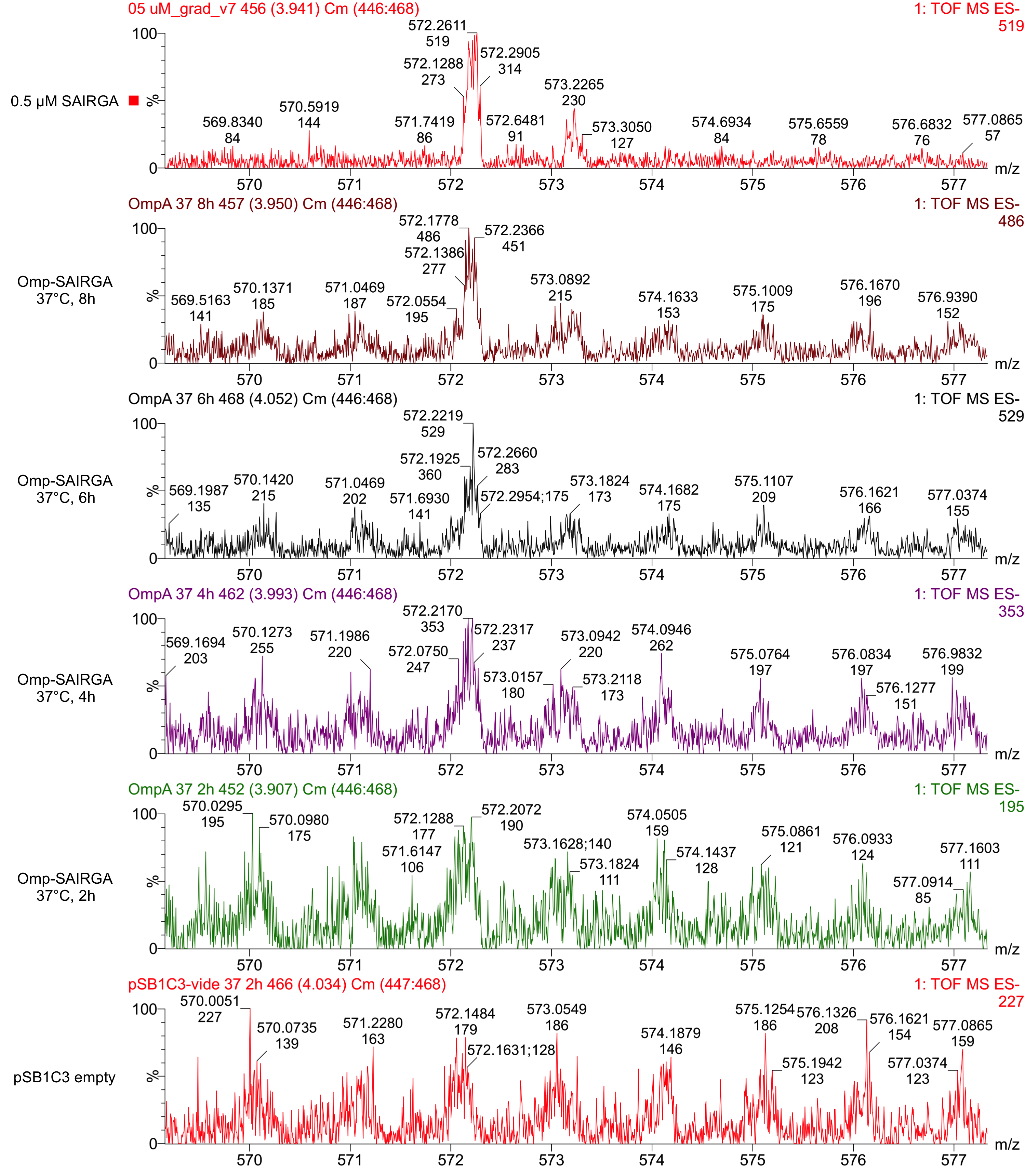
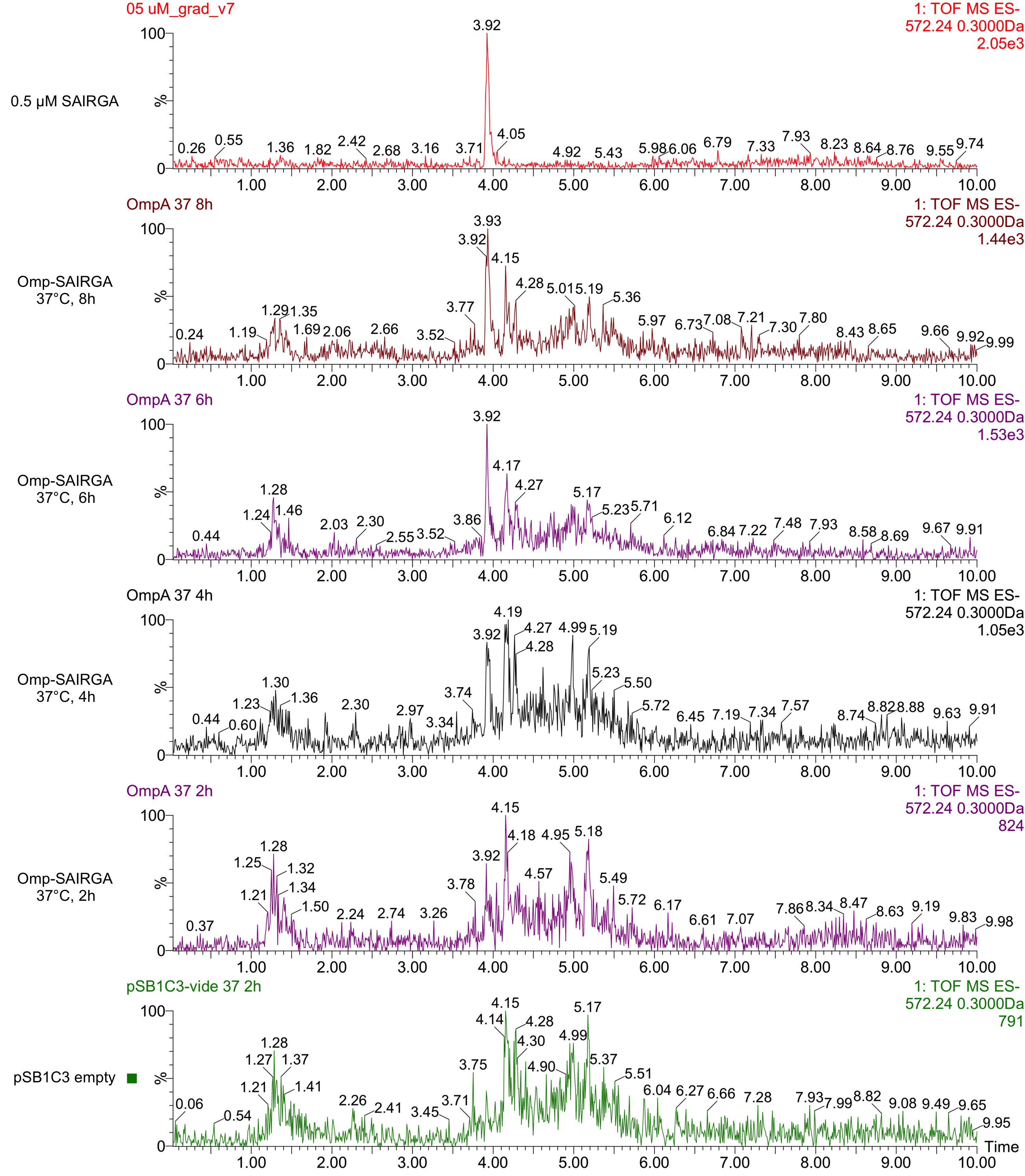
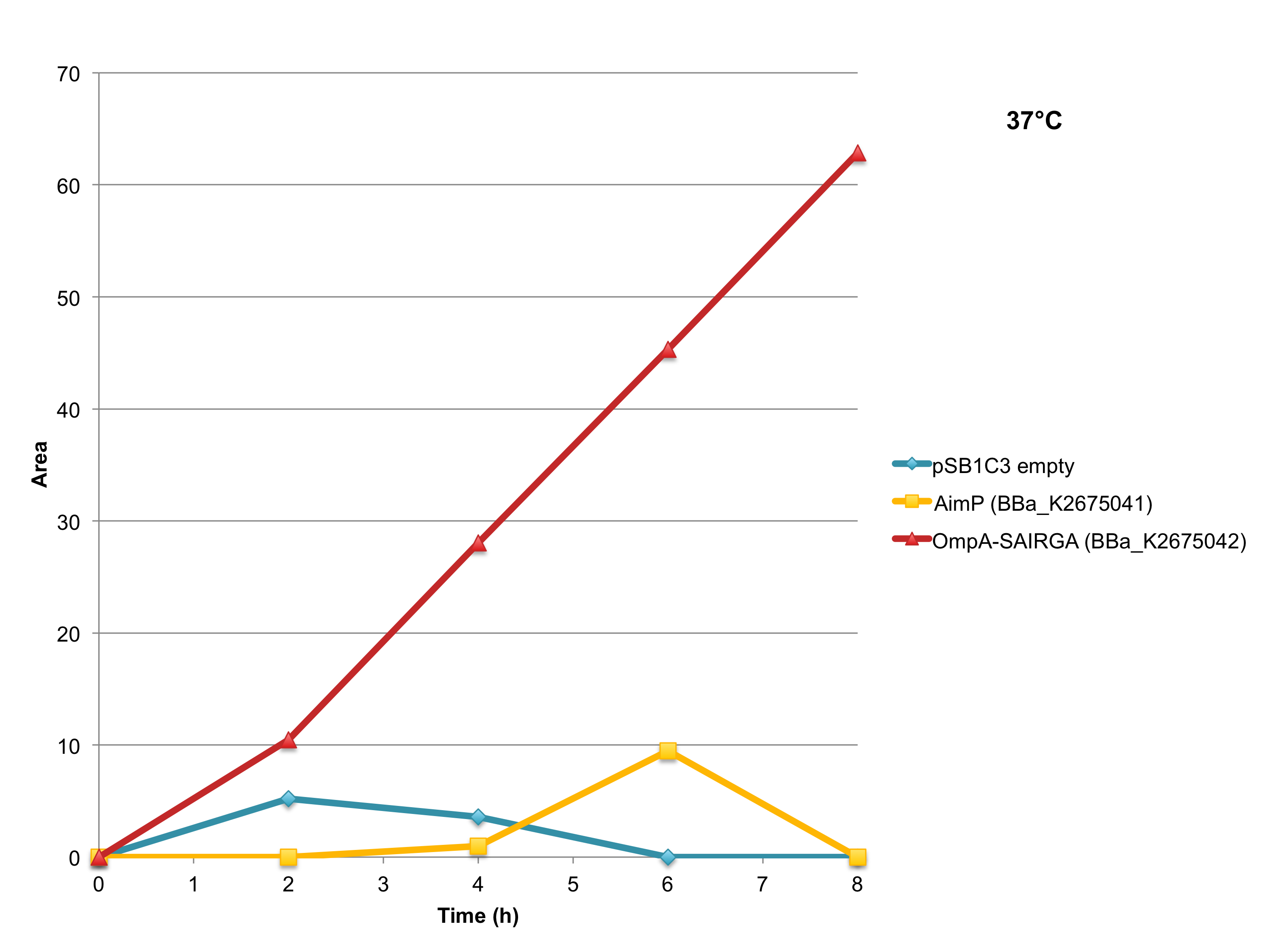
As shown in Figures 5 and 6, this expression vector of OmpA-SAIRGA is able to produce a compound with a monoisotopic mass a peak of 572 Da, which correspond to SAIRGA’s monoisotopic mass [M-H]- at a retention time of 3.9 min. The size or the area under the peak is increasing with time (figure 7). This suggests that OmpA is an efficient E. coli secretion tag which allows the release of the mature peptide SAIRGA in the culturing media.
Figure 5: Mass spectrometry of culturing media taken after 2, 4, 6 and 8 hours of incubation of E. coli cells harboring this OmpA-SAIRGA expression cassette in pSB1C3 (BBa_K2675042). The negative control has been performed with an empty pSB1C3 (bottom red chromatogram). The positive control (pure synthetic SAIRGA at 5 µM) is represented on the top red chromatogram.
Figure 6: UPLC chromatograms of culturing media taken after 2, 4, 6 and 8 hours of incubation of E. coli cells harboring this OmpA-SAIRGA expression cassette in pSB1C3 (BBa_K2675042). The negative control has been performed with an empty pSB1C3 (bottom green chromatogram). The positive control (pure synthetic SAIRGA at 5 µM) is represented on the top red chromatogram.
Figure 7: SAIRGA production at 37°C by E. coli cells harboring this OmpA-SAIRGA expression plasmid (BBa_K2675042) or BBa_K2675041 which is the AimP expression plasmid. The negative control has been performed with an empty pSB1C3.
References
[1] Erez Z, Steinberger-Levy I, Shamir M, Doron S, Stokar-Avihail A, Peleg Y, Melamed S, Leavitt A, Savidor A, Albeck S, Amitai G, Sorek R. Communication between viruses guides lysis-lysogeny decisions. Nature (2017) 541, 488-493.
[2] Espah Borujeni A, Channarasappa AS, Salis HM. Translation rate is controlled by coupled trade-offs between site accessibility, selective RNA unfolding and sliding at upstream standby sites. Nucleic Acids Res (2014) 42, 2646-2659.
[3] Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotechnol (2009) 27, 946-50.
[4] Hall BG. Activation of the bgl operon by adaptive mutation. Mol Biol Evol (1998) 15, 1-5.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]