Difference between revisions of "Part:BBa K2558001"
| Line 3: | Line 3: | ||
<partinfo>BBa_K2558001 short</partinfo> | <partinfo>BBa_K2558001 short</partinfo> | ||
| − | This part is derived from lux pR | + | This part is derived from lux pR. They are similar except for one point mutation in the luxR binding site (G19T) and another three in the -10 region of the promotor (TAG44CTT). The function remains the same: the mutant lux pR promotor is still inducible by LuxR-AHL complex. However, they do differ in gene expression strength, leakage and the expression parameters. |
===Usage and Biology=== | ===Usage and Biology=== | ||
Revision as of 12:27, 17 October 2018
lux pR-HS
This part is derived from lux pR. They are similar except for one point mutation in the luxR binding site (G19T) and another three in the -10 region of the promotor (TAG44CTT). The function remains the same: the mutant lux pR promotor is still inducible by LuxR-AHL complex. However, they do differ in gene expression strength, leakage and the expression parameters.
Usage and Biology
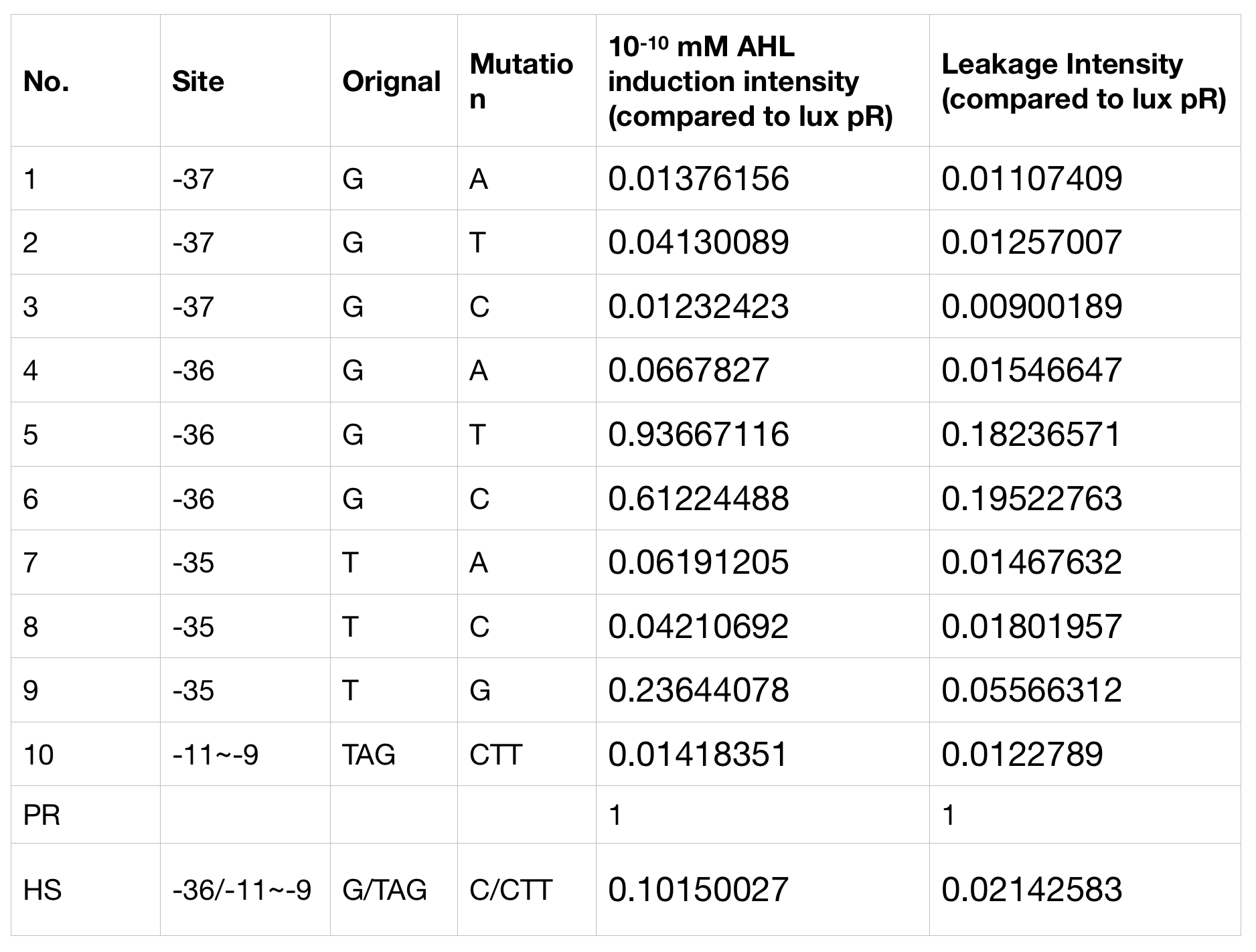
With the hope to find an optimal promotor for our NEON system, we designed 9 mutations on sites 18, 19 and 20 near the luxR binding site and tested one on -10 site that TUST 2017 concluded to show decreased leakage. We conducted experiments to evaluate these lux pR mutants’ reaction to AHL stimulation and their leakage level. The test devices we designed include a constantly expressed luxR and a lux pR (or mutant) (like BBa_K2558211 with original lux pR promotor https://parts.igem.org/Part:BBa_K2558211, and BBa_K2558212 with lux pR-HS promotor https://parts.igem.org/Part:BBa_K2558212).
Results
We conducted the experiment first time with 9 mutations on sites 18,19 and 20 near the luxR binding and 1 mutation on -10 site that TUST 2017 concluded to show decreased leakage. We have transferred the plasmids with lux pR mutants controlled sfGFP into E. coli. As we know, AHL could induce gene expression which is controlled by lux pR promoter. Therefore, we have measured the value of Fluorescence/OD without AHL by microplate reader to show the leakages of different lux pR mutants, as lux pR promoter is not supposed to be activated without AHL. Meanwhile, we also regard the value of Fluorescence/OD with 10-10 M AHL as the sensitivity of different lux pR mutants. The data of experiment is shown in the figures below. (Figure.1)
-
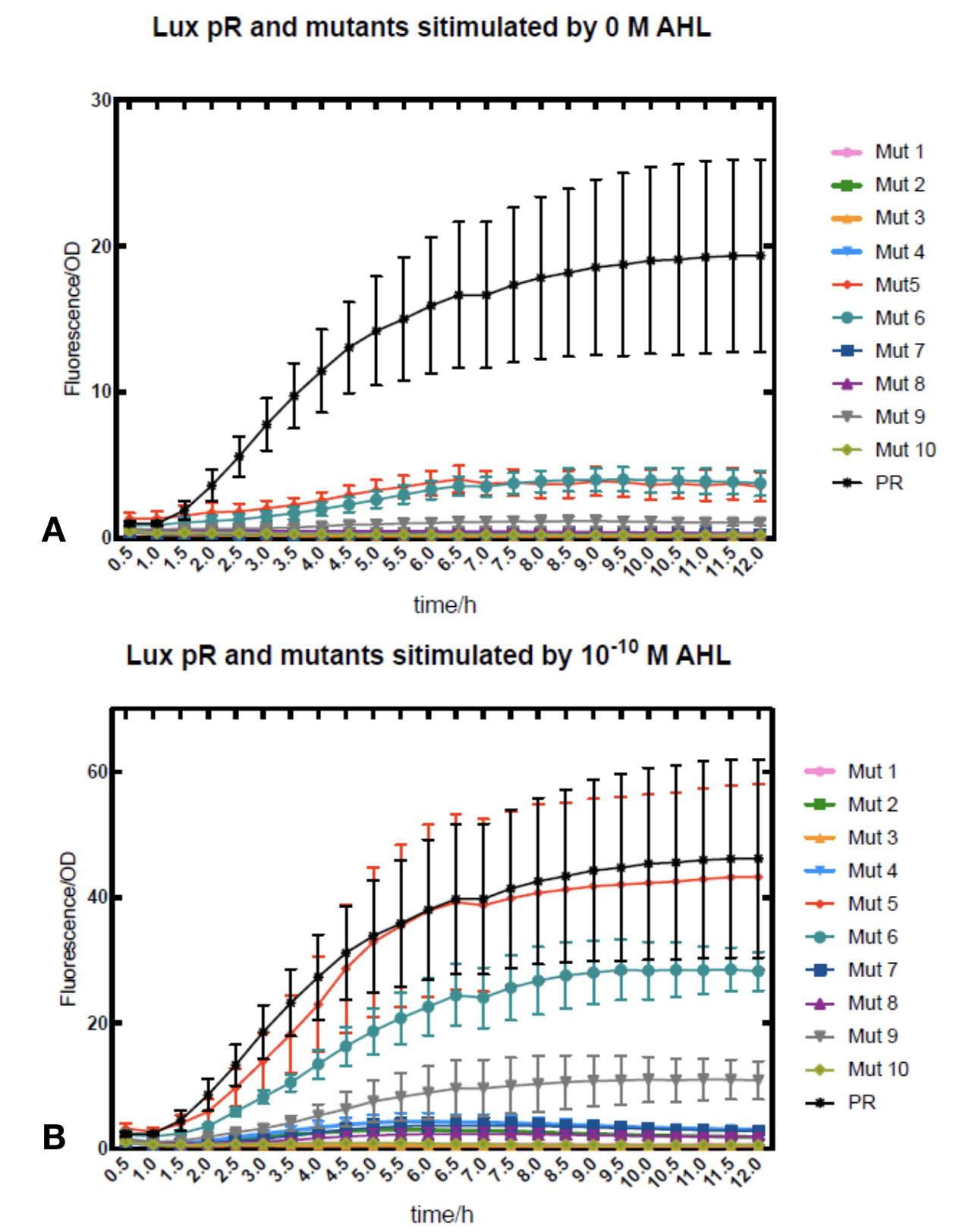 Figure.1. Leakage and 10-10 mM AHL stimulation of lux pR promoter and its mutations. The fluorescence strengths and OD values are measured by microplate reader at 510 nm and 600 nm wavelength. Plasmids with lux pR promoter and its mutants are transferred into E.coli DH5α. A The bacteria was treated without AHL inducing. The higher the ratio (Fluorescence/OD) is, the more leakage the promoter has. Among all the strains, the one with lux pR promoter has the largest leakage and the error bar as well. B The bacteria was treated with 10-10 mM AHL induction. The higher the ratio (Fluorescence/OD) is, the more sensitive the promoter is. Mutation 5 has almost the same sensitivity to the wild type lux pR promoter.
Figure.1. Leakage and 10-10 mM AHL stimulation of lux pR promoter and its mutations. The fluorescence strengths and OD values are measured by microplate reader at 510 nm and 600 nm wavelength. Plasmids with lux pR promoter and its mutants are transferred into E.coli DH5α. A The bacteria was treated without AHL inducing. The higher the ratio (Fluorescence/OD) is, the more leakage the promoter has. Among all the strains, the one with lux pR promoter has the largest leakage and the error bar as well. B The bacteria was treated with 10-10 mM AHL induction. The higher the ratio (Fluorescence/OD) is, the more sensitive the promoter is. Mutation 5 has almost the same sensitivity to the wild type lux pR promoter. -
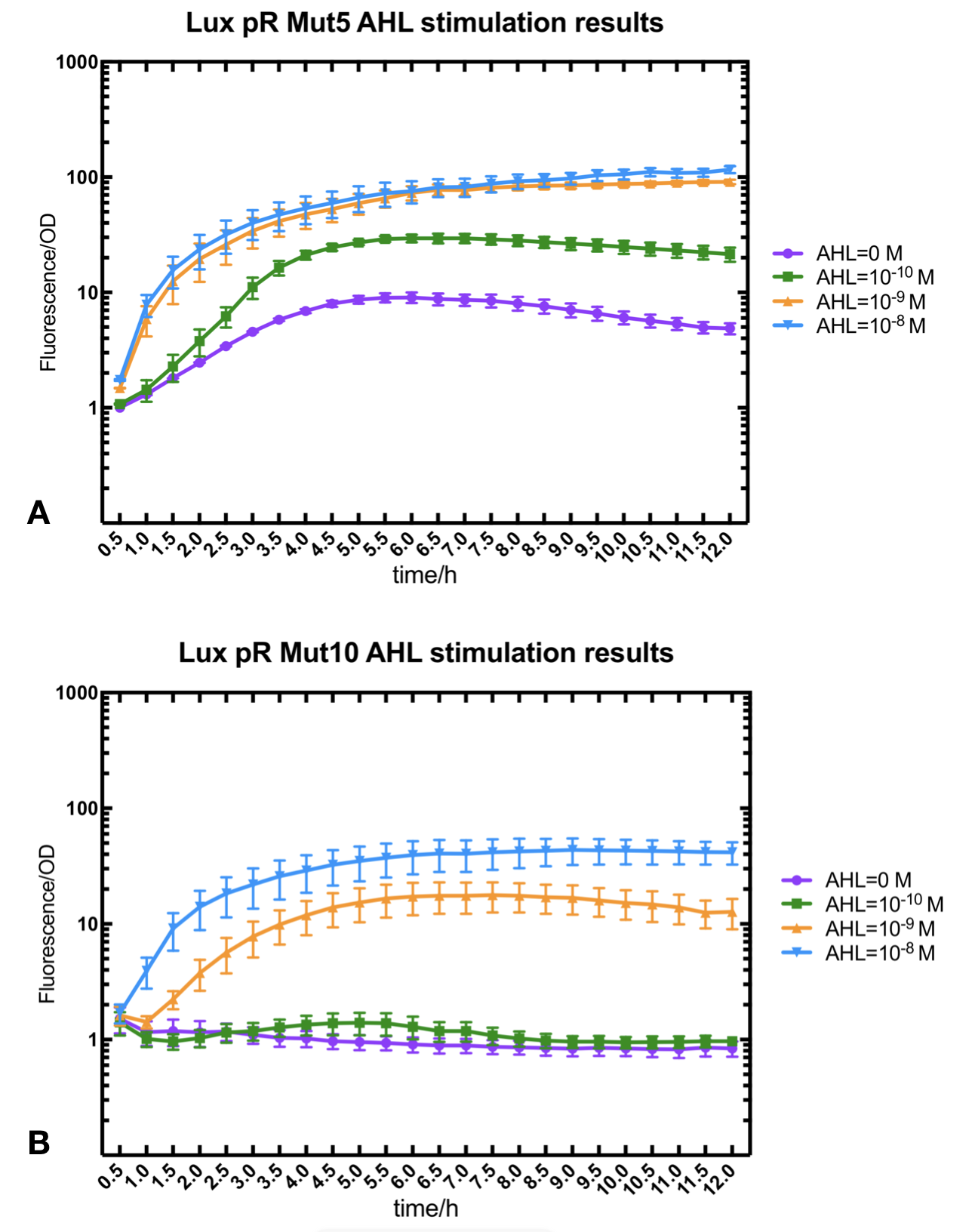 Figure.2. Leakage and AHL stimulation of of lux pR Mutant 5 and Mutant 10. The fluorescence strengths and OD values are measured by microplate reader at 510 nm and 600 nm wavelength. A Plasmid with lux pR Mutant 5 is transferred into E.coli DH5α. The bacteria was treated without or with 10-10 mM, 10-9 mM and 10-8 mM AHL inducing. B Plasmid with lux pR Mutant 10 is transferred into E.coli DH5α. The bacteria was treated without or with 10-10 mM, 10-9 mM and 10-8 mM AHL inducing.
Figure.2. Leakage and AHL stimulation of of lux pR Mutant 5 and Mutant 10. The fluorescence strengths and OD values are measured by microplate reader at 510 nm and 600 nm wavelength. A Plasmid with lux pR Mutant 5 is transferred into E.coli DH5α. The bacteria was treated without or with 10-10 mM, 10-9 mM and 10-8 mM AHL inducing. B Plasmid with lux pR Mutant 10 is transferred into E.coli DH5α. The bacteria was treated without or with 10-10 mM, 10-9 mM and 10-8 mM AHL inducing. -
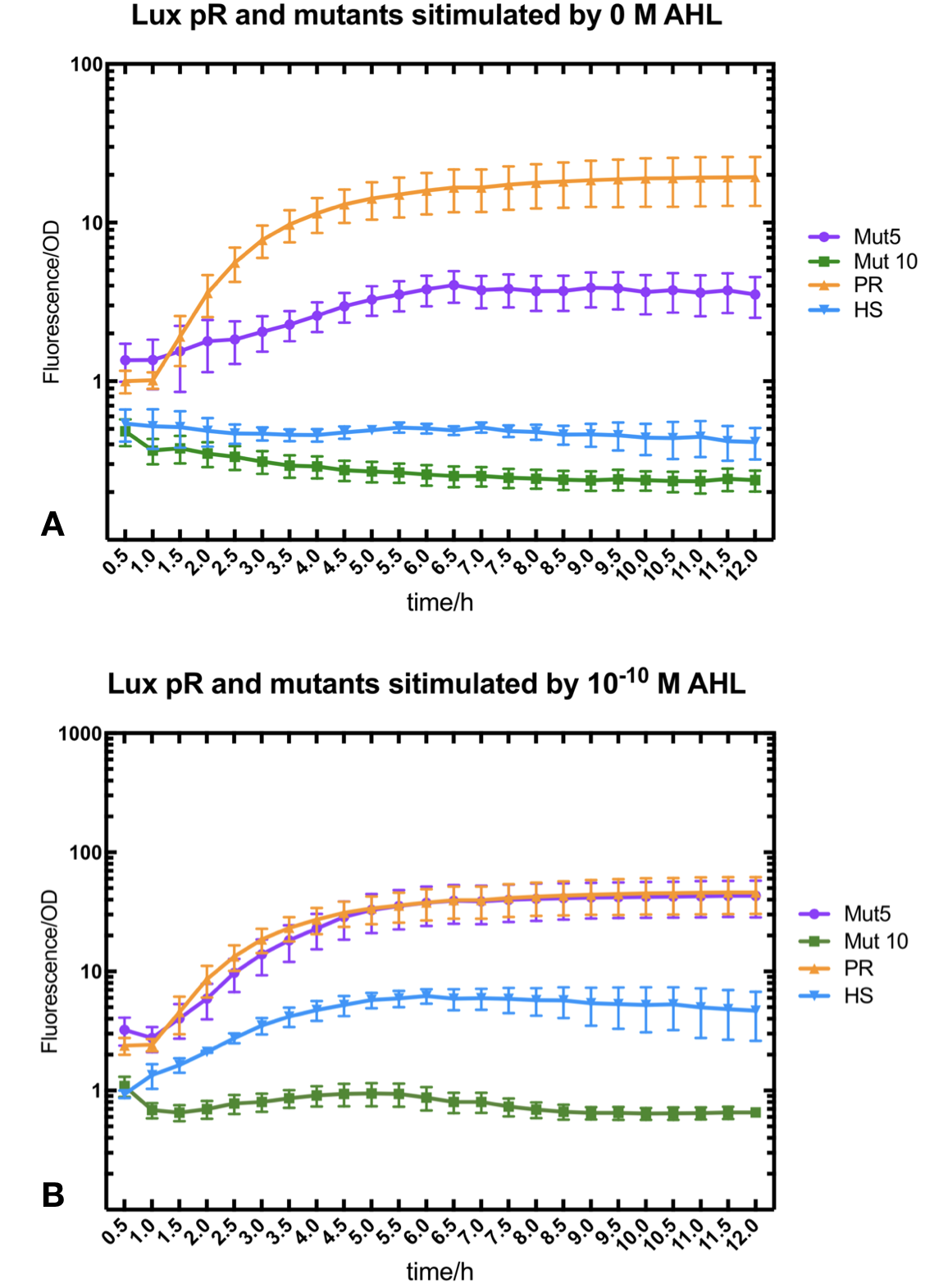 Figure.3. Leakage and 10^-10 mM AHL stimulation of of lux pR promoter WT, Mutant 5, Mutant 10 and HS. The fluorescence strengths and OD values are measured by microplate reader at 510 nm and 600 nm wavelength. Plasmids with lux pR promoter WT, Mutant 5 and Mutant 10 are transferred into E.coli DH5α. A The bacteria was treated without AHL inducing. The higher the ratio (Fluorescence/OD) is, the more leakage the promoter has. B The bacteria was treated with 10-10 mM AHL induction. The higher the ratio (Fluorescence/OD) is, the more sensitive the promoter is.
Figure.3. Leakage and 10^-10 mM AHL stimulation of of lux pR promoter WT, Mutant 5, Mutant 10 and HS. The fluorescence strengths and OD values are measured by microplate reader at 510 nm and 600 nm wavelength. Plasmids with lux pR promoter WT, Mutant 5 and Mutant 10 are transferred into E.coli DH5α. A The bacteria was treated without AHL inducing. The higher the ratio (Fluorescence/OD) is, the more leakage the promoter has. B The bacteria was treated with 10-10 mM AHL induction. The higher the ratio (Fluorescence/OD) is, the more sensitive the promoter is.It is observed that the higher leakage lux pR mutant has, the more sensitive this promoter mutation is. The wild type of lux pR shown with black strand in figures has the highest level of both leakage and sensibility. The next three are Mutant 5 (G-36T), Mutant 6 (G-36C), Mutant 9 (T-35G). Mutant 5 has almost the same sensitivity to wild type, but the leakage is much smaller than the wild type. Mutant 6 and Mutant 9 has a little bit less leakage and medium sensitivity. We have also done the experiment with 10-9 M and 10-8 M AHL, the all promoters show high expression levels. (Data not shown) We repeated AHL stimulation test on Mutant 5 and Mutant 10 (Figure.2) and decided to combine the two mutations together to form a new promoter with high sensitive and small leakage. We named it lux pR-HS. We tested lux pR-HS under the same conditions as those mutations. We can see that the lux pR-HS has similar leakage to Mutant 10, while it has much higher sensitivity. (Figure.3) The statistics of relative induction and leakage intensity of the 10 Mutants and lux pR-HS are shown in Table.1
-
As for why the mutation happened on -35 to -37 sites would decrease the leakage of lux pR without severely influencing the expression level or sensibility, we have some hypothesis and explanations. It is reported that -35 to -37 sites are last three bases of lux box which is upstream of the lux promoter and bound specifically by luxR, one of regulatory protein involved in lux expression system.[1] G19T mutation (-36 G to T mutation upstream of lux pR promoter) has less influence than other mutation on sites -35 to -37, which is same to our result. Besides, the G19T is not involved in the two important regions of nucleotides 3 to 5 and 16 to 18 which directly contacted with luxR protein. Therefore, this single change of nucleotide would decrease the leakage of lux promoter without influencing the expression level too much.[2]
Protocol
- one Transform the plasmids into E. coli DH5α.
- two Pick a single colony by a sterile tip from each of the LB plates for all the experimental and control groups. Add the colony into 5ml LB medium with ampicillin at 100 ng/µl. Incubate for 6-8 h at 37℃ in a shaker.
- three Measure OD600 of the culture medium with photometer. Dilute the culture medium until OD600 reaches 0.6.
- four Add 100 µl bacteria culture medium into a sterile 96-well plate. Add AHL to final concentrations of 0, 10-10,10-9,10-8 M. Fresh LB medium serves as blank control. Positive control is colony constantly expressing sfGFP and negative control is colony without sfGFP expression. Place the 96-well plate into an automatic microplate reader. Incubate at 16℃ overnight and record the fluorometric value at 510 nm and OD600 for each well every 30 minutes.
- five Each group is repeated for at least 3 times.
Reference
[1] Antunes, L. C., et al. "A mutational analysis defines Vibrio fischeri LuxR binding sites." Journal of Bacteriology 190.13(2008):4392-4397.
[2] Zeng, Weiqian, et al. "Rational design of an ultrasensitive quorum-sensing switch." Acs Synthetic Biology 6.8(2017).
Sequence and Features
Assembly Compatibility:- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]