Difference between revisions of "Part:BBa K1330000"
Frederickk (Talk | contribs) |
Frederickk (Talk | contribs) |
||
| Line 39: | Line 39: | ||
===2018 Team Hong_Kong-CUHK's Improvement=== | ===2018 Team Hong_Kong-CUHK's Improvement=== | ||
Spinach2.1 was originally constructed to prevent the illegal site in the superfolding Spinach2. However, its activity might be moderately less bright. On the other hand, Spinach2 has the melting temperature of ~38 degrees Celsius according to previous literature, which might not favor the measurement of heat-induced promoter activity. | Spinach2.1 was originally constructed to prevent the illegal site in the superfolding Spinach2. However, its activity might be moderately less bright. On the other hand, Spinach2 has the melting temperature of ~38 degrees Celsius according to previous literature, which might not favor the measurement of heat-induced promoter activity. | ||
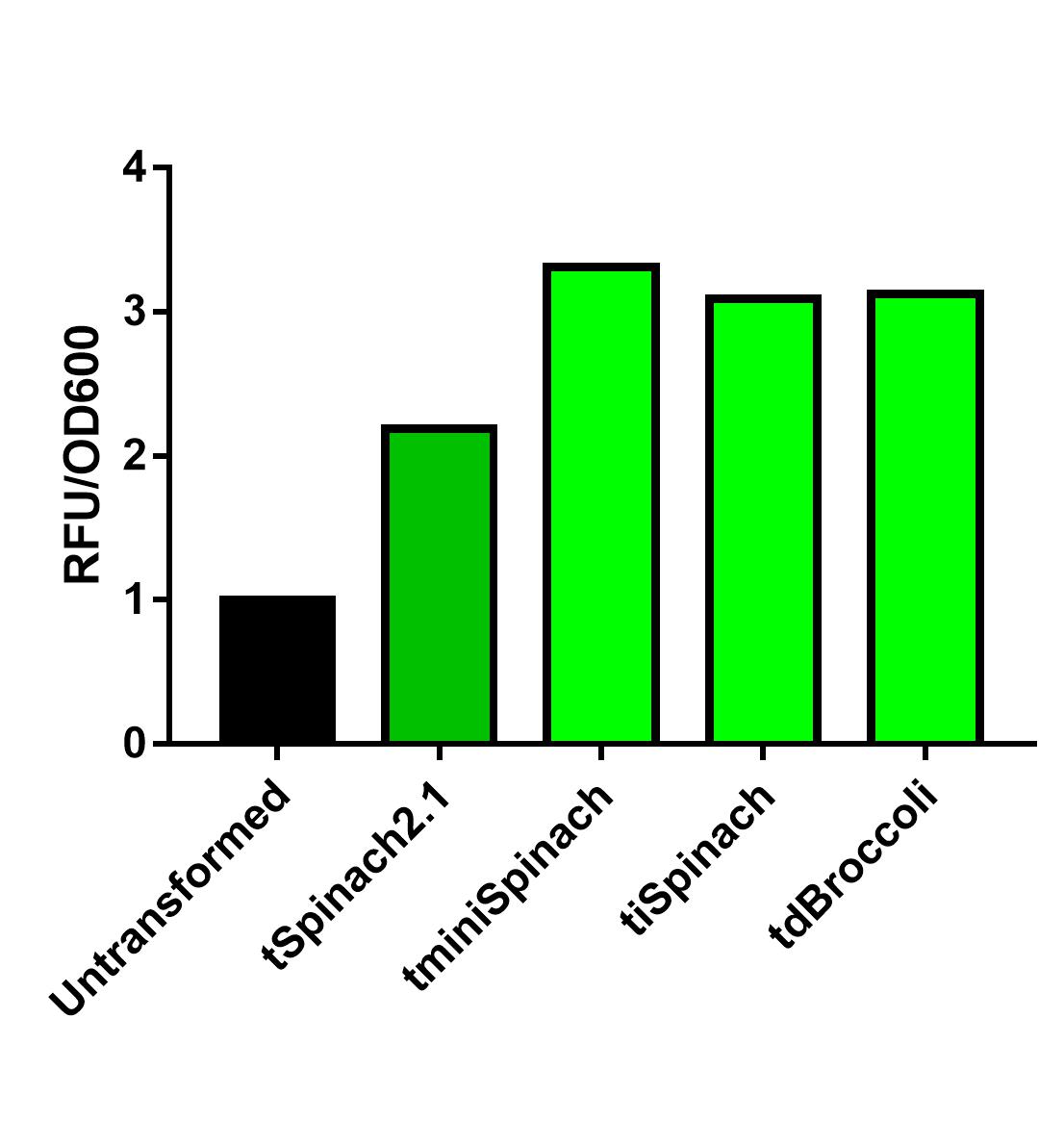
| + | Comparing the 3 RNA reporters we constructed and the pre-existed Spinach2.1, iSpinach-D5 is moderately brighter. | ||
| + | [[File:Improve a previous bobrick.jpg|400px|thumb|left|Fluorescence data obtained from plate reader. DH5a transformed with pSB1C3-lpp-Spinach2.1 or other Spinach variants were incubated overnight at 37 degrees Celsius. Overnight culture was normalized to OD=0.2 and incubated in 200uM DFHBI for 45 minutes. N=1. More replicates are needed.]] | ||
| + | <p style="clear:left"></p> | ||
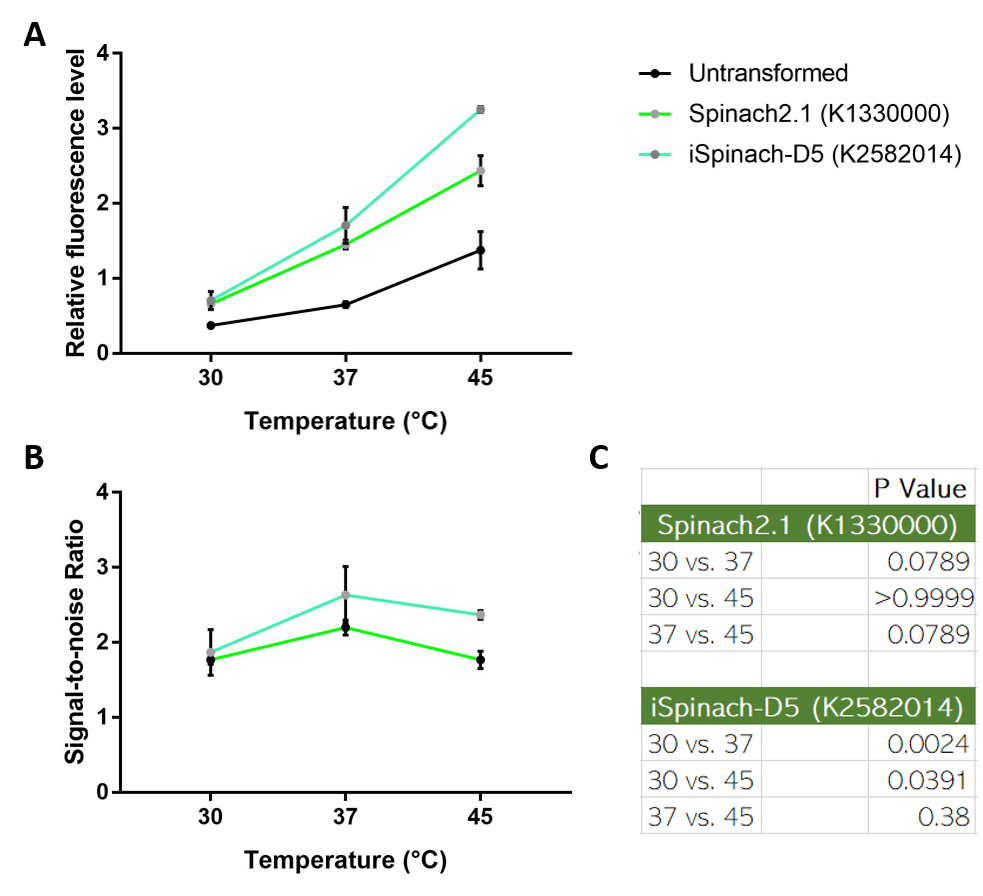
| + | Collaborating with NUS-A team, we also observed that iSpinach-D5 is moderately more resistant to 37->45 degrees Celsius change than Spinach2.1. However, it was also more vulnerable to 37->30 degrees Celsius change. | ||
| + | [[File:ImproveNUS.png|700px|thumb|left|(A) Fluorescence data obtained from plate reader. DH5a transformed with pSB1C3-lpp-Spinach2.1 or iSpinach-D5 were incubated overnight at 30, 37 or 45 degrees Celsius. Overnight culture was normalized to OD=0.2 and incubated in 200uM DFHBI for 45 minutes at their incubation temperatures. (B) Signal-to-noise ratio calculated by dividing fluorescence level of Spinach-expressing E. coli with that of untransformed E. coli. (C) Multiple t-tests across temperatures. 37 vs 45 P-value of iSpinach is larger than that of Spinach2.1. N=3.]] | ||
| + | <p style="clear:left"></p> | ||
<!-- --> | <!-- --> | ||
Revision as of 19:43, 16 October 2018
Spinach2.1 flanked by tRNALys3
This BioBrick contains the gene coding for the Spinach2.1 RNA, flanked by a tRNA scaffold sequence.
Usage and Biology
Spinach 2.1 RNA can bind and activate the fluorophore [http://pubs.acs.org/doi/abs/10.1021/ja410819x DFHBI-1T], both in vivo and in vitro. RNA molecules can thus be fluorescently tagged, by gene fusion with this BioBrick. The flanking tRNA comes from human tRNALys3 and increases stability and folding efficiency of the RNA, leading to higher fluorescent signal. It is possible to detect signal from Spinach2.1 when expressed from moderate strength promoters, e.g. the Anderson promoter library.
This part does not contain or need a Ribosome Binding Site.
Characterization
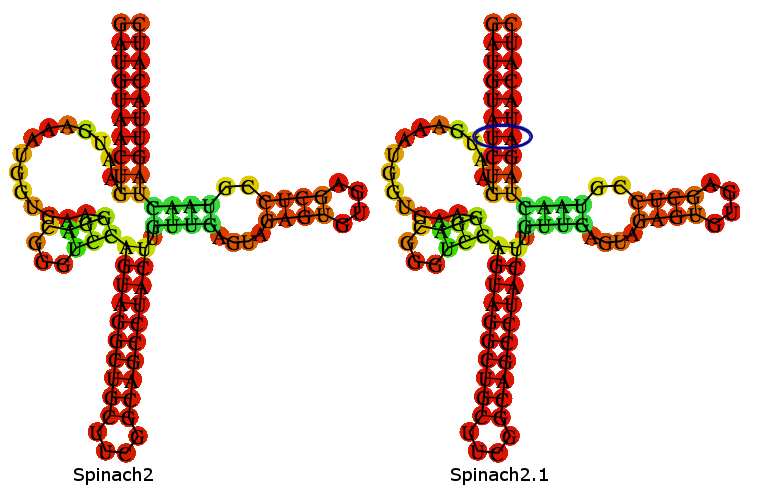
Spinach2.1 is derived from the Spinach2 sequence described in [http://www.nature.com/nmeth/journal/v10/n12/full/nmeth.2701.html Strack et al. (2013)]. Spinach2.1 has had 2 nucleotides swapped to remove a SpeI site, present in Spinach2.
A folding-prediction is shown below, demonstrating that Spinach2.1 is expected adopt the same fold as Spinach2.
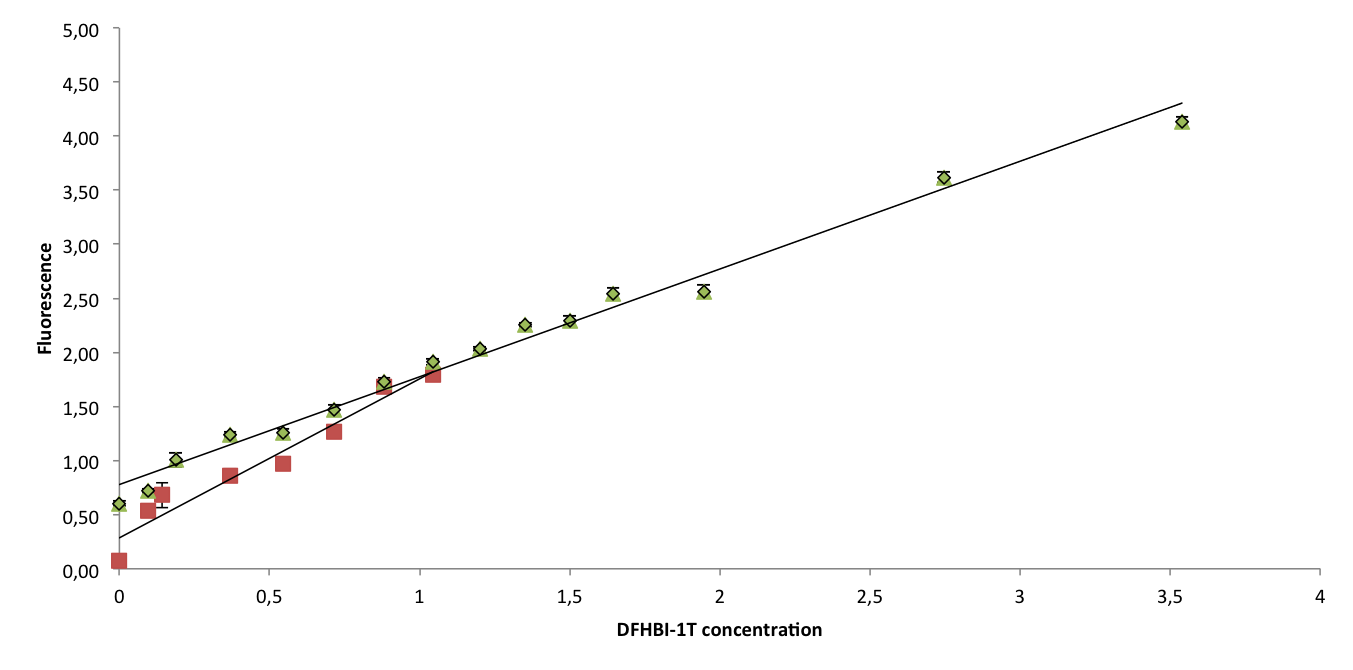
To confirm that Spinach2.1 functions as well as Spinach2, fluorescence measurements were conducted on Spinach2 and Spinach2.1 RNA in vitro. Fluorescence was measured on samples with different concentrations of DFHBI-1T and excess RNA, to compare the brightness of the Spinach-DFHBI-1T complexes. The resulting fluorescence standard curves are shown below:
The slopes of the curves represent the brightness of the RNA-DFHBI-1T complex. The slopes are similar, although Spinach2.1 might be moderately less bright. More experiments need to be conducted to establish whether the difference is significant.
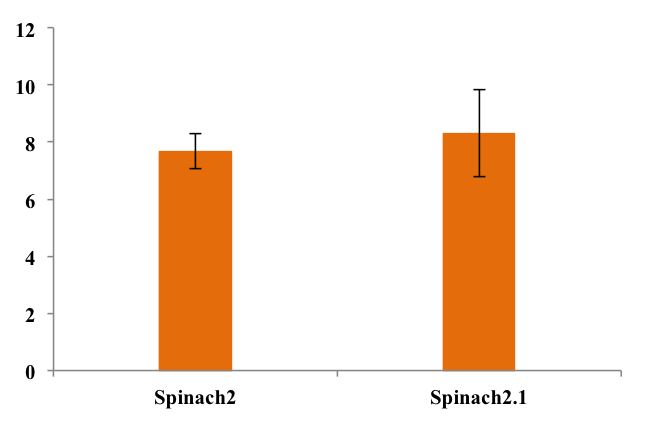
To assess folding efficiency of Spinach2.1 fluorescence was measured on samples with excess DFHBI-1T. Fluorescence normalized to RNA concentration is shown below:
As seen from the chart, there was no significant difference between fluorescence from the two RNA's. This means that equal fractions of RNA was bound to DFHBI-1T, indicating equal folding efficiency.
See [http://2014.igem.org/Team:DTU-Denmark/Achievements/Experimental_Results#lab-comparison-div our wiki] for more details.
2018 Team Hong_Kong-CUHK's Improvement
Spinach2.1 was originally constructed to prevent the illegal site in the superfolding Spinach2. However, its activity might be moderately less bright. On the other hand, Spinach2 has the melting temperature of ~38 degrees Celsius according to previous literature, which might not favor the measurement of heat-induced promoter activity. Comparing the 3 RNA reporters we constructed and the pre-existed Spinach2.1, iSpinach-D5 is moderately brighter.
Collaborating with NUS-A team, we also observed that iSpinach-D5 is moderately more resistant to 37->45 degrees Celsius change than Spinach2.1. However, it was also more vulnerable to 37->30 degrees Celsius change.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]