Difference between revisions of "Part:BBa K2728003"
Bluepumpkin (Talk | contribs) |
Bluepumpkin (Talk | contribs) (→Origin) |
||
| Line 19: | Line 19: | ||
=== Origin === | === Origin === | ||
| − | Nanoluc luciferase is a commercial product from PROMEGA | + | Nanoluc luciferase is a commercial product from PROMEGA. It was engineered by directed evolution from a deep-sea shrimp (Oplophorus gracilirostris) luciferase. |
<br /> | <br /> | ||
<br /> | <br /> | ||
| + | |||
=== Experimental Characterization === | === Experimental Characterization === | ||
We used the original plasmid provided by Promega to verify its luminescence strength. We transferred the plasmid into E. coli BL21 strain, a common bacterial strain for protein expression. 200 μl of the overnight bacteria was added into the 96-well plate for the luminescence assay. The luminescence curve was tested under the condition of emission wavelength=465nM and orbital shaking with amplitude=2. We can observed from the result that | We used the original plasmid provided by Promega to verify its luminescence strength. We transferred the plasmid into E. coli BL21 strain, a common bacterial strain for protein expression. 200 μl of the overnight bacteria was added into the 96-well plate for the luminescence assay. The luminescence curve was tested under the condition of emission wavelength=465nM and orbital shaking with amplitude=2. We can observed from the result that | ||
Revision as of 11:29, 14 October 2018
NanoLuc
Basic Description
NanoLuc (Nluc) luciferase is a small enzyme (19.1kDa) engineered for optimal performance as a luminescent reporter. The enzyme is about 100-fold brighter than either firefly (Photinus pyralis) or Renilla reniformis luciferase using a novel substrate, furimazine, to produce high intensity, glow-type luminescence. The luminescent reaction is ATP-independent and designed to suppress background luminescence for maximal assay sensitivity.
Sequence and Features
NanoLuc luciferase possesses a number of physical properties that make it an excellent reporter protein:
- very small, monomeric enzyme (171 amino acids; 513bp)
- high thermal stability (Tm = 60°C)
- active over a broad pH range (pH 6–8)
- no post-translational modifications or disulfide bonds
- uniform distribution in cells
- emission spectrum well suited for bioluminescence resonance energy transfer (BRET; λmax = 465nM)
Origin
Nanoluc luciferase is a commercial product from PROMEGA. It was engineered by directed evolution from a deep-sea shrimp (Oplophorus gracilirostris) luciferase.
Experimental Characterization
We used the original plasmid provided by Promega to verify its luminescence strength. We transferred the plasmid into E. coli BL21 strain, a common bacterial strain for protein expression. 200 μl of the overnight bacteria was added into the 96-well plate for the luminescence assay. The luminescence curve was tested under the condition of emission wavelength=465nM and orbital shaking with amplitude=2. We can observed from the result that
Potential Application
The nanoluc luciferase can be used as a reporter of formaldehyde. We can calculate the formaldehyde concentration according to the strength of luminescence and the model of luminescence-formaldehyde concentration. What’s more, it can be used in the product so that customers can judge the existence of formaldehyde by observing the luminescence.
References
Goyet, E., et al., Fast and high resolution single-cell BRET imaging. Sci Rep, 2016. 6: p. 28231.
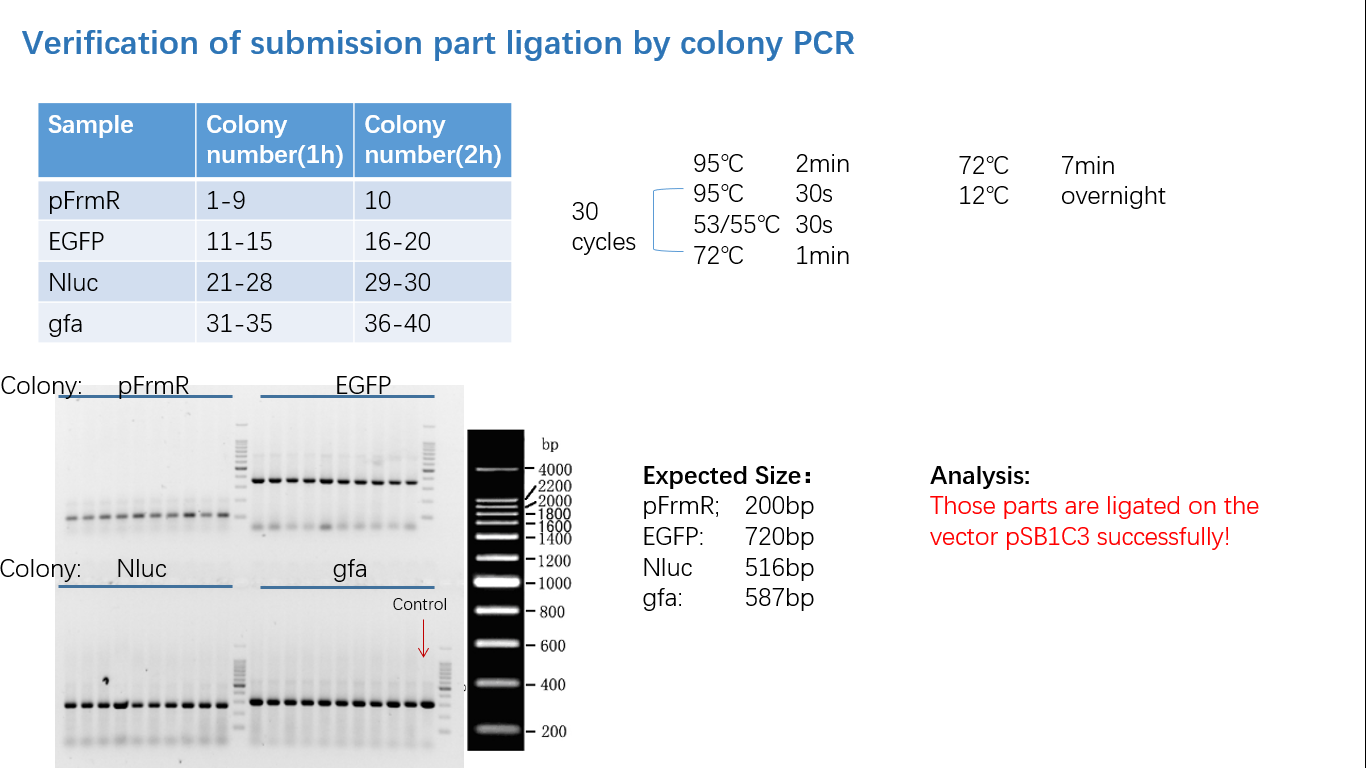
Parts Verification Before Submission
We verified our parts in the lab before submission. They are reliable! Please feel free to apply them onto your project.=)
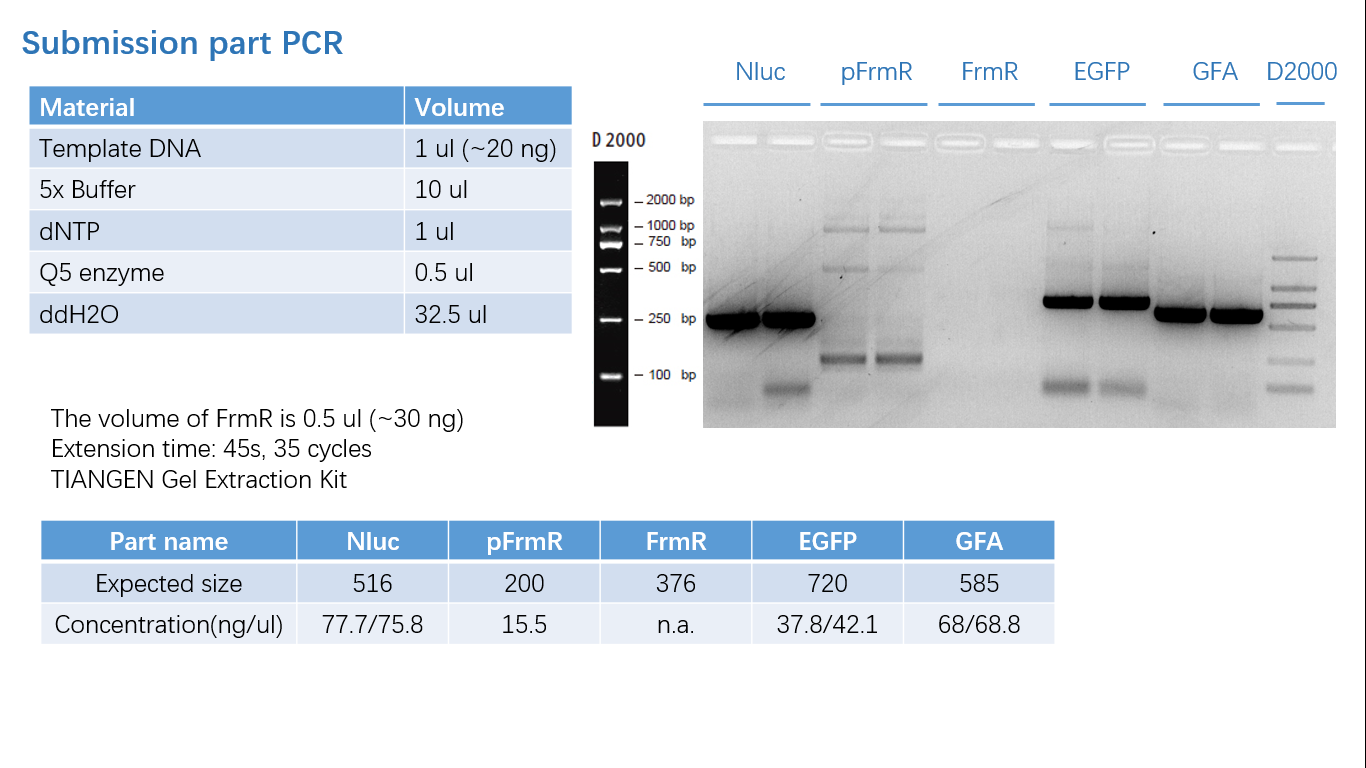
Fig 1: PCR (to get targeted genes)
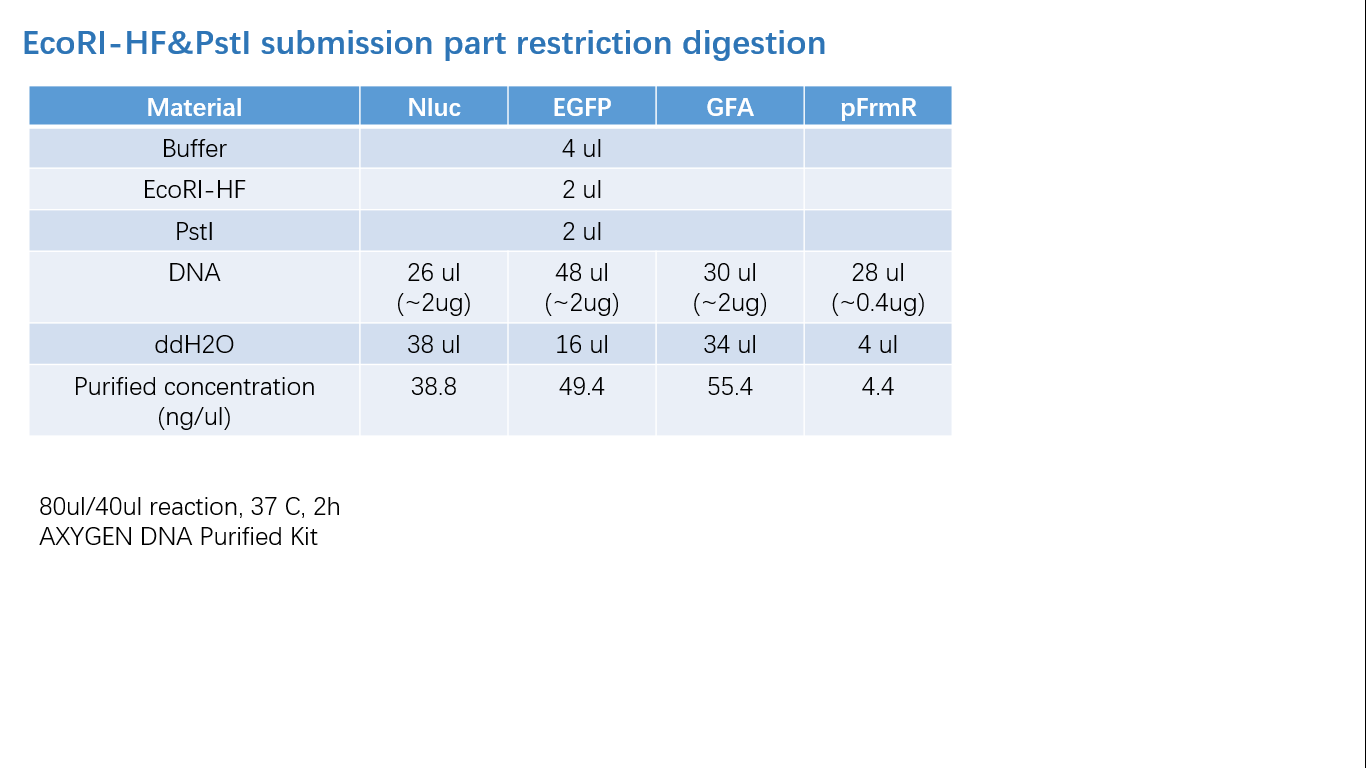
Fig 2: Restriction Digestion
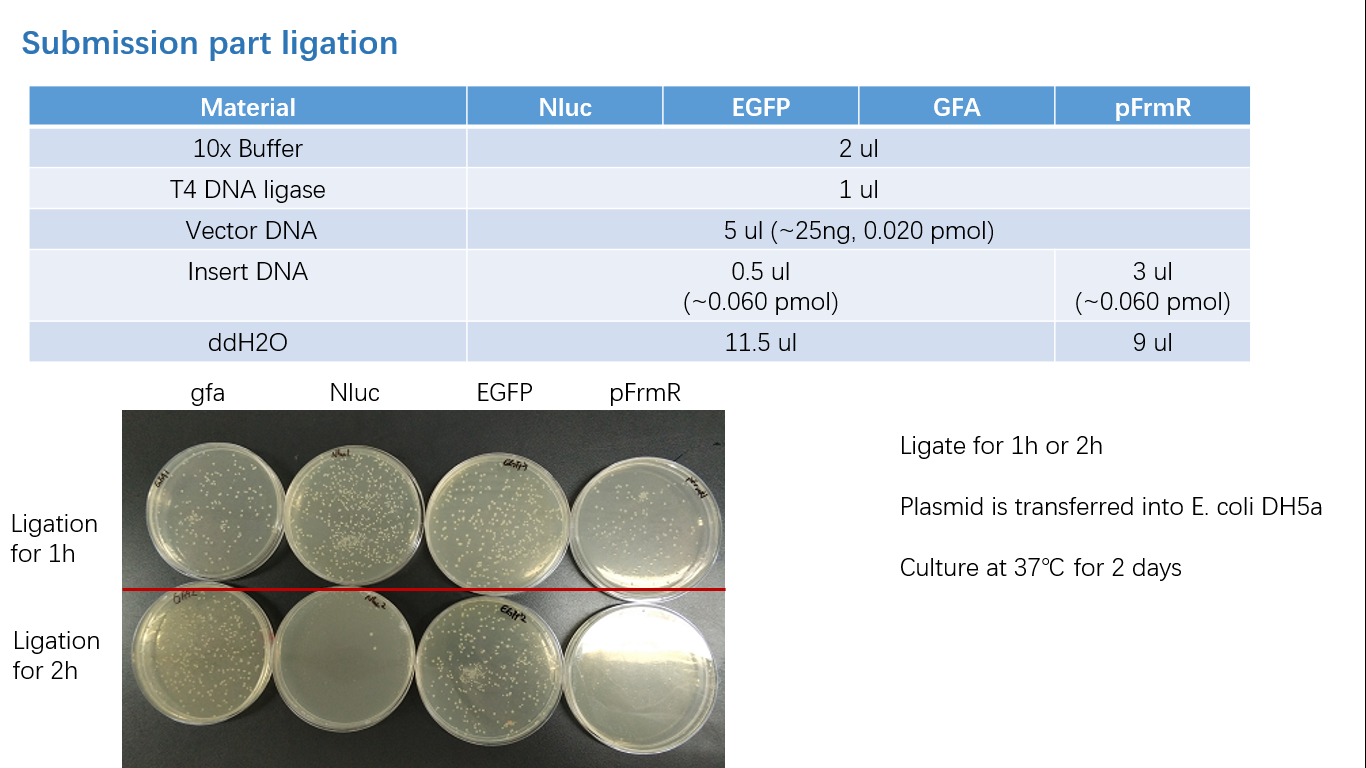
Fig 3: Ligation
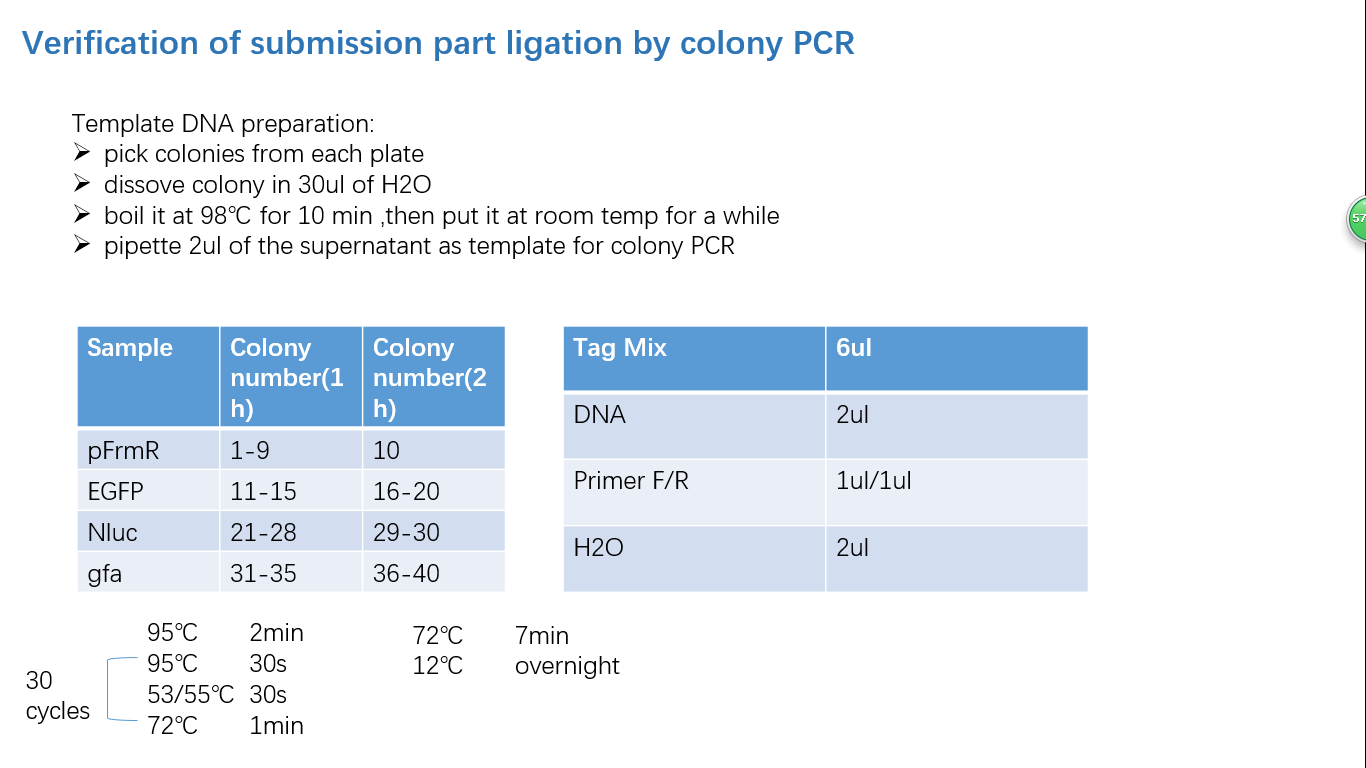
Fig 4: Colony PCR
Fig 5: Gel Verification
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 46
- 1000COMPATIBLE WITH RFC[1000]