Difference between revisions of "Part:BBa K2728004"
Bluepumpkin (Talk | contribs) |
Bluepumpkin (Talk | contribs) |
||
| Line 2: | Line 2: | ||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K2728004 short</partinfo> | <partinfo>BBa_K2728004 short</partinfo> | ||
| − | + | <br /> | |
| + | <br /> | ||
=== Basic Description === | === Basic Description === | ||
An enzyme from Paracoccus denitrificans that accelerates this spontaneous condensation reaction, which catalyze the conversion of formaldehyde and glutathione was purified and named glutathione-dependent formaldehyde-activating enzyme (Gfa). | An enzyme from Paracoccus denitrificans that accelerates this spontaneous condensation reaction, which catalyze the conversion of formaldehyde and glutathione was purified and named glutathione-dependent formaldehyde-activating enzyme (Gfa). | ||
| Line 14: | Line 15: | ||
===== Fig 2: 3D structure (from www.uniprot.org) ===== | ===== Fig 2: 3D structure (from www.uniprot.org) ===== | ||
<br /> | <br /> | ||
| − | + | <br /> | |
=== Sequence === | === Sequence === | ||
We ordered the synthesized plasmid from Genscript. After restriction digestion & transformation, we got our E. coli with GFA. | We ordered the synthesized plasmid from Genscript. After restriction digestion & transformation, we got our E. coli with GFA. | ||
| Line 22: | Line 23: | ||
===== Fig 3: Order info ===== | ===== Fig 3: Order info ===== | ||
<br /> | <br /> | ||
| − | + | <br /> | |
=== Origin === | === Origin === | ||
This gene originated from P. denitrificans. Putative proteins with sequence identity to GFA from P. denitrificans are present also in Rhodobacter sphaeroides, Sinorhizobium meliloti, and Mesorhizobium loti. | This gene originated from P. denitrificans. Putative proteins with sequence identity to GFA from P. denitrificans are present also in Rhodobacter sphaeroides, Sinorhizobium meliloti, and Mesorhizobium loti. | ||
<br /> | <br /> | ||
| − | + | <br /> | |
=== Experimental Characterization === | === Experimental Characterization === | ||
We have tested the influence of GFA in different formaldehyde concentration. Cultures were inoculated from overnights to an optical density (OD) of 0.05 in 5 mL in 15 mL disposable culture tubes, incubated at 37 °C for 2−3 h, and dosed with 0−500 μM formaldehyde (MP Biomedicals, Santa Ana, CA). And we aimed at comparing the effect of GFA test E. coli with normal E. coli BL21 in formaldehyde environment by OD600nm. But in 100~500uM formaldehyde, there are no difference in BL21 with test E. coli. | We have tested the influence of GFA in different formaldehyde concentration. Cultures were inoculated from overnights to an optical density (OD) of 0.05 in 5 mL in 15 mL disposable culture tubes, incubated at 37 °C for 2−3 h, and dosed with 0−500 μM formaldehyde (MP Biomedicals, Santa Ana, CA). And we aimed at comparing the effect of GFA test E. coli with normal E. coli BL21 in formaldehyde environment by OD600nm. But in 100~500uM formaldehyde, there are no difference in BL21 with test E. coli. | ||
<br /> | <br /> | ||
| − | + | <br /> | |
=== References === | === References === | ||
Goenrich, M., Bartoschek, S., Hagemeier, C. H., Griesinger, C. & Vorholt, J. A. (2002). A glutathione-dependent formaldehydeactivating enzyme (Gfa) from Paracoccus denitrificans detected and purified via two-dimensional proton exchange NMR spectroscopy.J Biol Chem 277, 3069–3072. | Goenrich, M., Bartoschek, S., Hagemeier, C. H., Griesinger, C. & Vorholt, J. A. (2002). A glutathione-dependent formaldehydeactivating enzyme (Gfa) from Paracoccus denitrificans detected and purified via two-dimensional proton exchange NMR spectroscopy.J Biol Chem 277, 3069–3072. | ||
Rohlhill J, Sandoval N R, Papoutsakis E T. Sort-seq approach to engineering a formaldehyde-inducible promoter for dynamically regulated Escherichia coli growth on methanol.[J]. Acs Synthetic Biology, 2017, 6(8). | Rohlhill J, Sandoval N R, Papoutsakis E T. Sort-seq approach to engineering a formaldehyde-inducible promoter for dynamically regulated Escherichia coli growth on methanol.[J]. Acs Synthetic Biology, 2017, 6(8). | ||
| + | <br /> | ||
| + | <br /> | ||
| + | === Parts Verification Before Submission === | ||
| + | We verified our parts in the lab before submission. They are reliable! Please feel free to apply them onto your project.=) | ||
| + | [[File:T--BGIC-Global--partsub1.png|left|border|800px]]<br clear=all> | ||
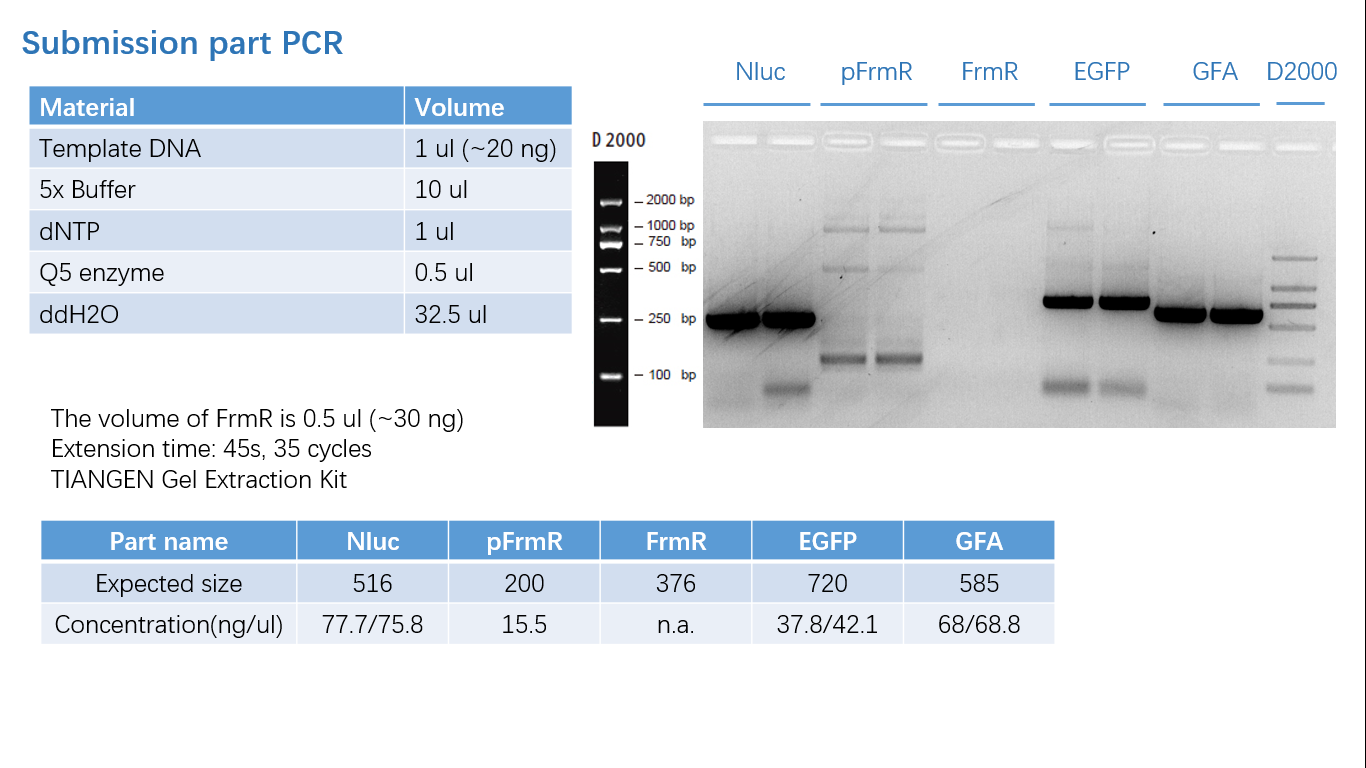
| + | ===== Fig 1: PCR (to get targeted genes) ===== | ||
| + | <br /> | ||
| + | [[File:T--BGIC-Global--partsub2.png|left|border|800px]]<br clear=all> | ||
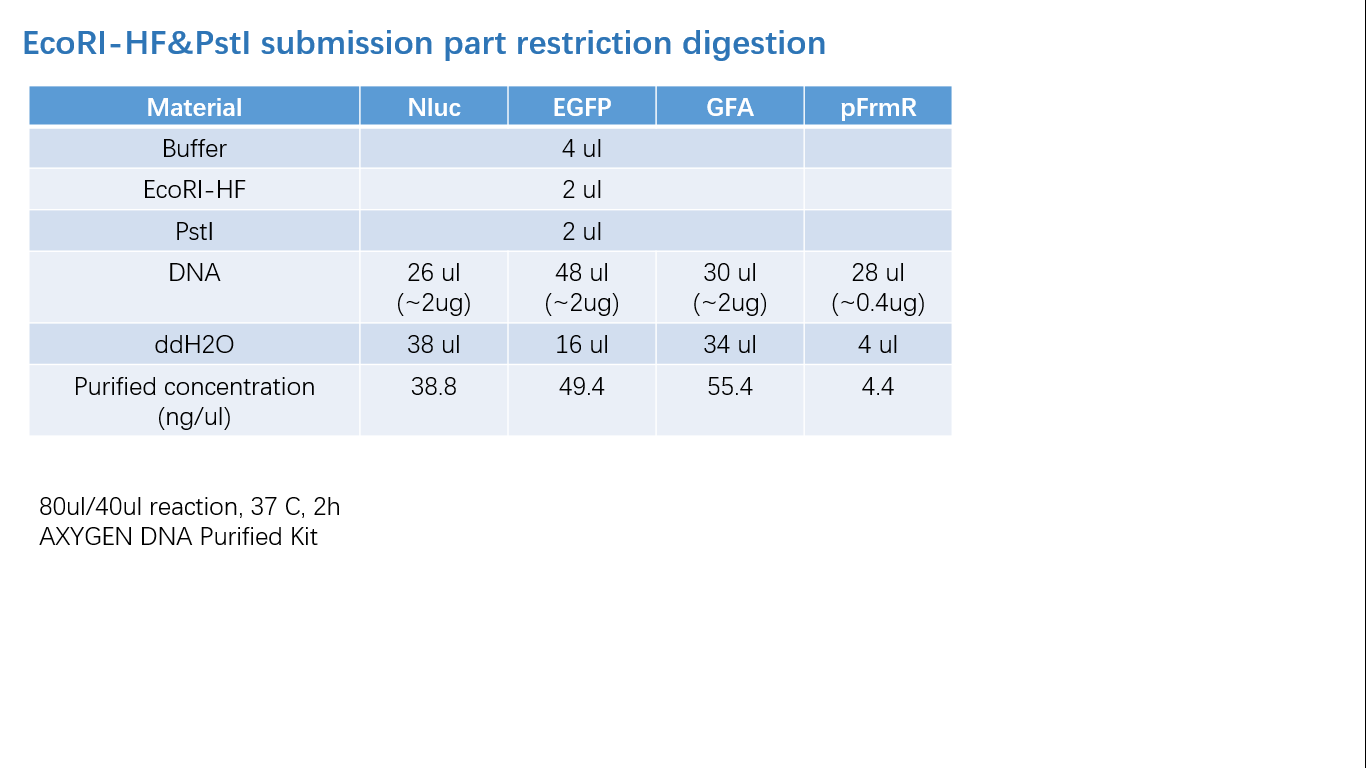
| + | ===== Fig 2: Restriction Digestion ===== | ||
| + | <br /> | ||
| + | [[File:T--BGIC-Global--partsub3.png|left|border|800px]]<br clear=all> | ||
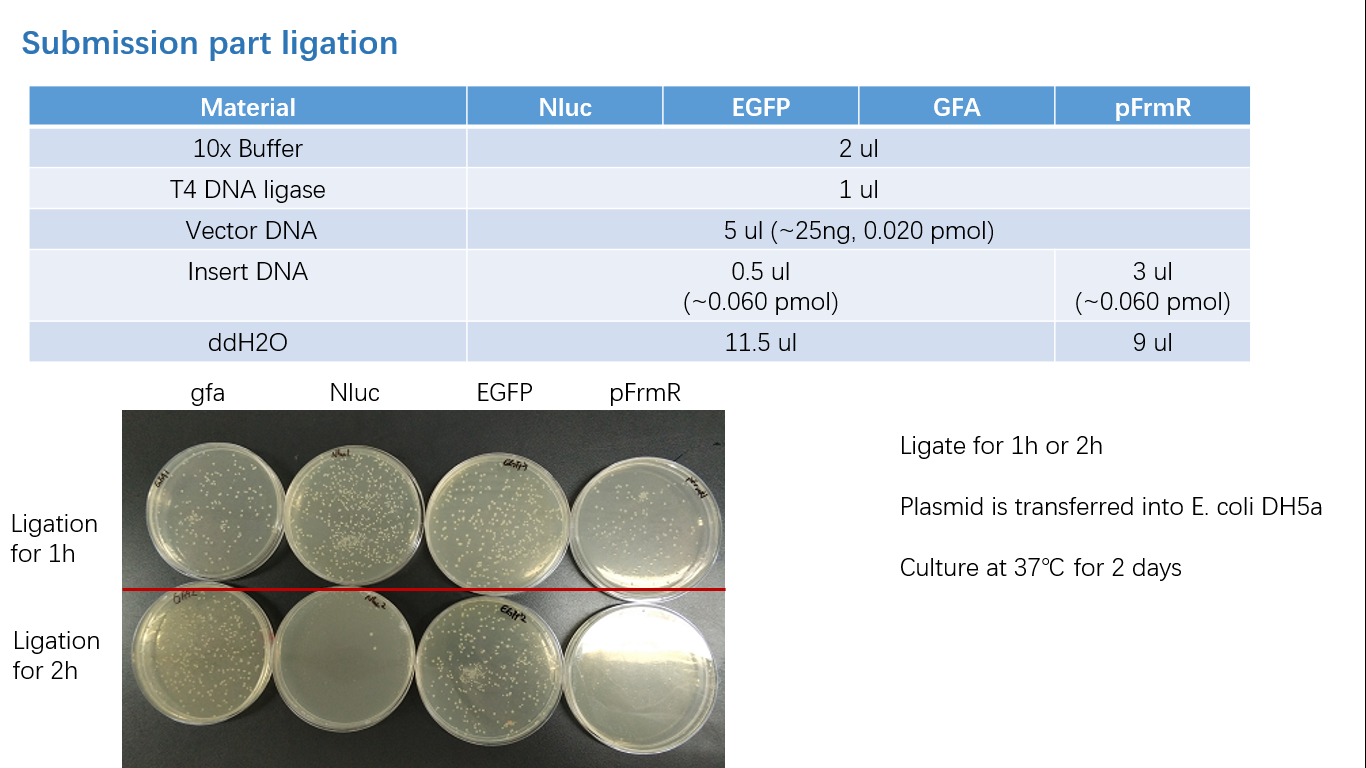
| + | ===== Fig 3: Ligation ===== | ||
| + | <br /> | ||
| + | [[File:T--BGIC-Global--partsub4.png|left|border|800px]]<br clear=all> | ||
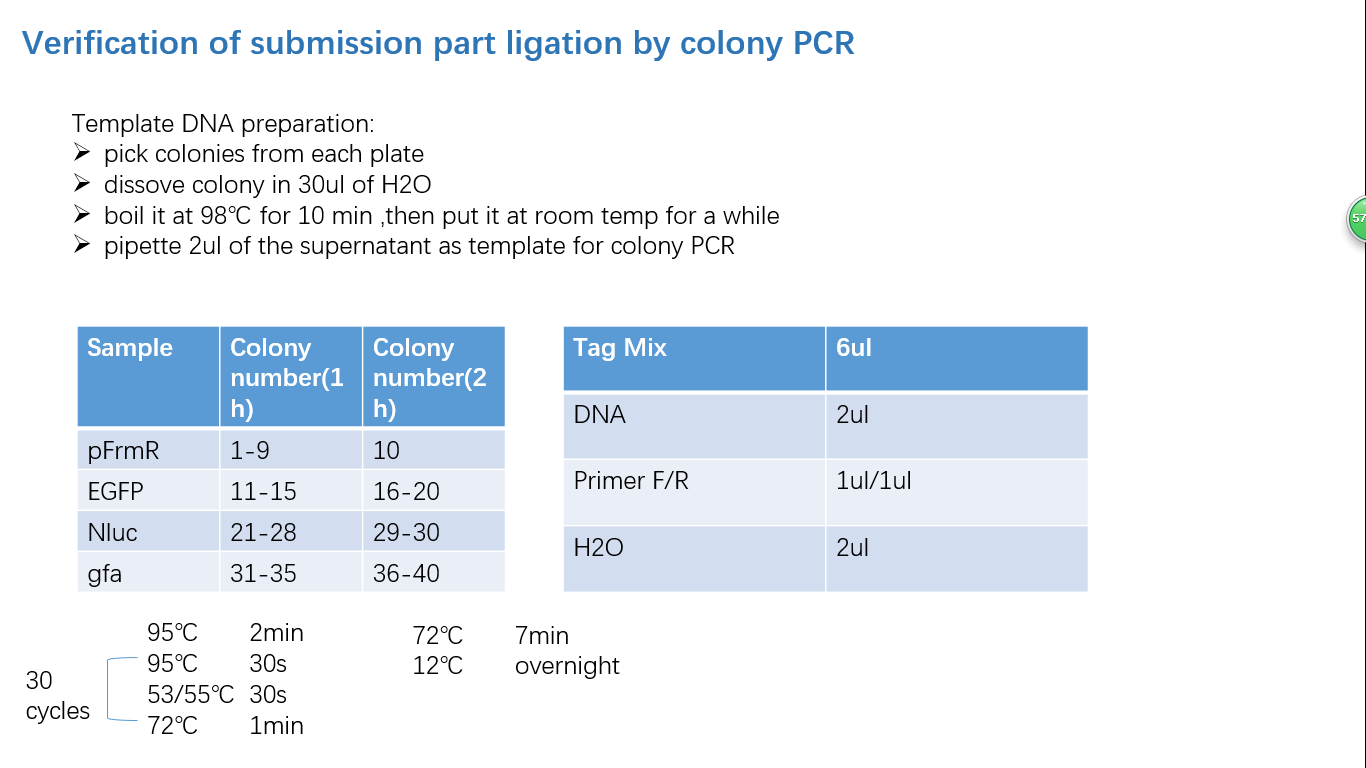
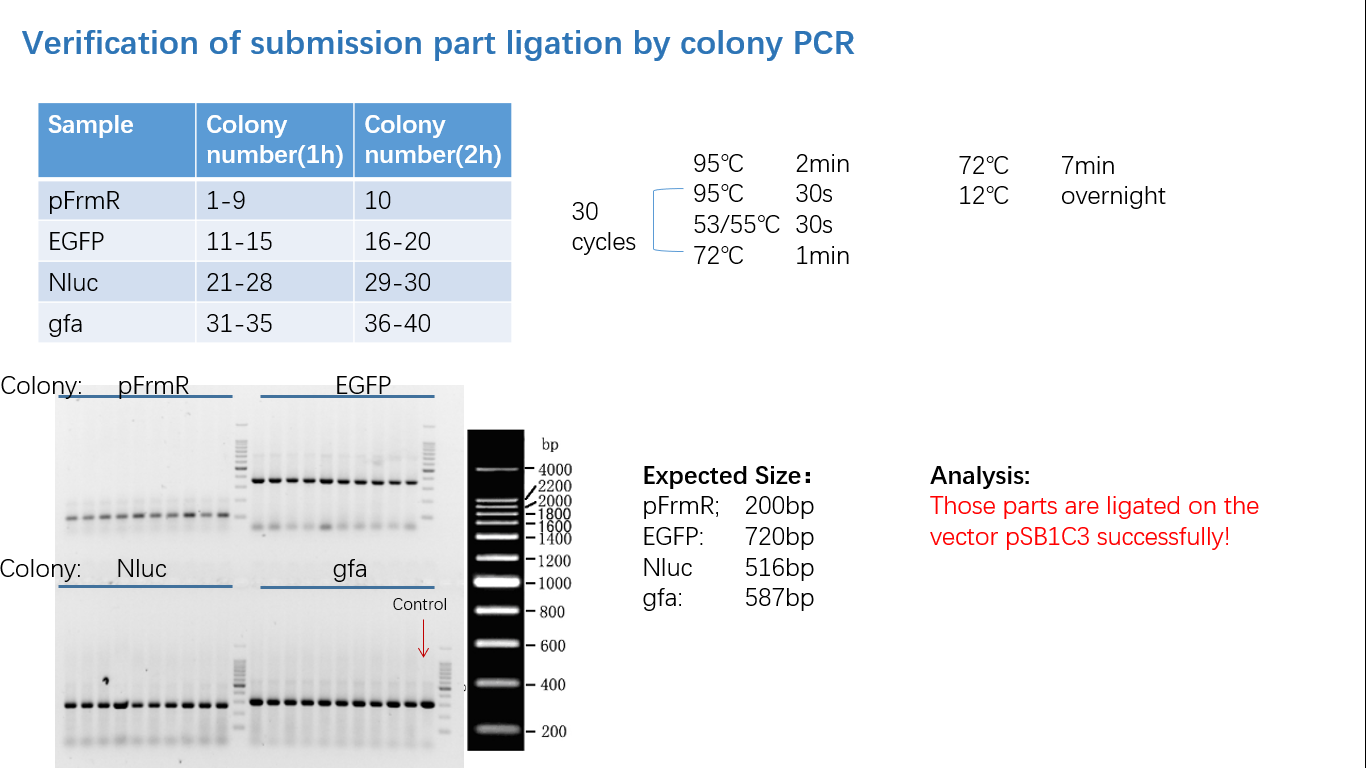
| + | ===== Fig 4: Colony PCR ===== | ||
| + | <br /> | ||
| + | [[File:T--BGIC-Global--partsub5.png|left|border|800px]]<br clear=all> | ||
| + | ===== Fig 5: Gel Verification ===== | ||
<br /> | <br /> | ||
Revision as of 04:37, 14 October 2018
GFA - A Glutathione-Dependent Formaldehyde-Activating Enzyme
Basic Description
An enzyme from Paracoccus denitrificans that accelerates this spontaneous condensation reaction, which catalyze the conversion of formaldehyde and glutathione was purified and named glutathione-dependent formaldehyde-activating enzyme (Gfa).
The gene GFA is located directly upstream of the gene for glutathione-dependent formaldehyde dehydrogenase, which catalyzes the subsequent oxidation of S-hydroxymethylglutathione. The glutathione-dependent formaldehyde conversion to formate starts with the adduct formation, formaldehyde reacts with the SH group of glutathione producing S-hydroxymethylglutathione (Reaction 1).
Fig 1: Thiol-dependent pathway
Formaldehyde-converting enzymes-Gfa is composed of one type of subunit of about 20 kDa and lack a chromophoric prosthetic group. In addition, both enzymes are encoded next to genes for enzymes involved in further oxidation of the cofactor-bound one-carbon unit to carbon dioxide.
Fig 2: 3D structure (from www.uniprot.org)
Sequence
We ordered the synthesized plasmid from Genscript. After restriction digestion & transformation, we got our E. coli with GFA.
Fig 3: Order info
Origin
This gene originated from P. denitrificans. Putative proteins with sequence identity to GFA from P. denitrificans are present also in Rhodobacter sphaeroides, Sinorhizobium meliloti, and Mesorhizobium loti.
Experimental Characterization
We have tested the influence of GFA in different formaldehyde concentration. Cultures were inoculated from overnights to an optical density (OD) of 0.05 in 5 mL in 15 mL disposable culture tubes, incubated at 37 °C for 2−3 h, and dosed with 0−500 μM formaldehyde (MP Biomedicals, Santa Ana, CA). And we aimed at comparing the effect of GFA test E. coli with normal E. coli BL21 in formaldehyde environment by OD600nm. But in 100~500uM formaldehyde, there are no difference in BL21 with test E. coli.
References
Goenrich, M., Bartoschek, S., Hagemeier, C. H., Griesinger, C. & Vorholt, J. A. (2002). A glutathione-dependent formaldehydeactivating enzyme (Gfa) from Paracoccus denitrificans detected and purified via two-dimensional proton exchange NMR spectroscopy.J Biol Chem 277, 3069–3072.
Rohlhill J, Sandoval N R, Papoutsakis E T. Sort-seq approach to engineering a formaldehyde-inducible promoter for dynamically regulated Escherichia coli growth on methanol.[J]. Acs Synthetic Biology, 2017, 6(8).
Parts Verification Before Submission
We verified our parts in the lab before submission. They are reliable! Please feel free to apply them onto your project.=)
Fig 1: PCR (to get targeted genes)
Fig 2: Restriction Digestion
Fig 3: Ligation
Fig 4: Colony PCR
Fig 5: Gel Verification
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]