Difference between revisions of "Part:BBa K2601002"
LebronJames (Talk | contribs) |
LebronJames (Talk | contribs) |
||
| Line 10: | Line 10: | ||
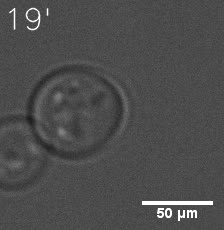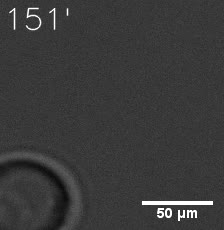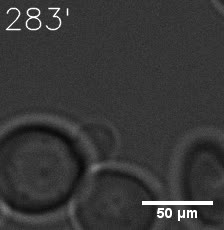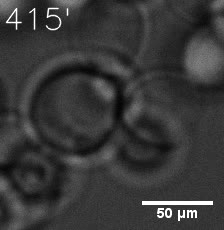
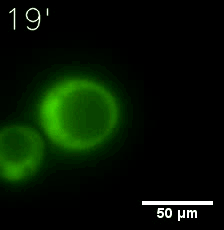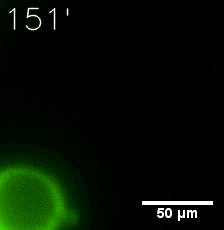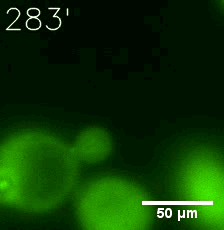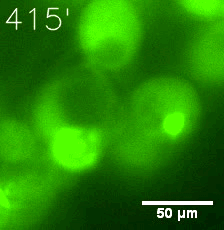
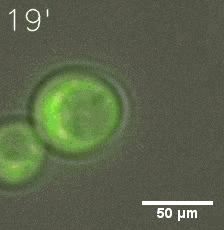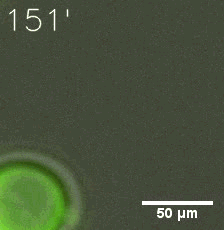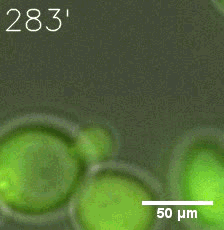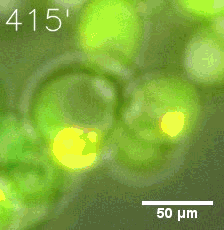
Some membrane-less organelles, such as stress granules and P bodies, have been discovered in recent years. Proteins condense into droplets and assemble these organelles through a process called phase separation. | Some membrane-less organelles, such as stress granules and P bodies, have been discovered in recent years. Proteins condense into droplets and assemble these organelles through a process called phase separation. | ||
| − | [[file:T--Peking--phase_separation_demo1.png|250px]] | + | <table border="0" align="center"> <tr> <td>[[file:T--Peking--phase_separation_demo1.png|250px]]</td> <td>[[file:T--Peking--phase_separation_demo2.png|250px]</td> </tr> |
In order to rationally design a synthetic organelle based on protein phase separation, we need a multivalent module and a protein-protein interaction module. The paired SUMO and SIM is one of the bioparts that we chose to introduce protein-protein interaction. SUMO and SIM can dimerize spontaneously. | In order to rationally design a synthetic organelle based on protein phase separation, we need a multivalent module and a protein-protein interaction module. The paired SUMO and SIM is one of the bioparts that we chose to introduce protein-protein interaction. SUMO and SIM can dimerize spontaneously. | ||
Revision as of 05:31, 11 October 2018
SUMO (Small Ubiquitin-like Modifier)
Introduction
SUMOylation is a post-translational modification, which is involved in many cellular processes and provide a versatile way to regulate the dynamics of protein-protein interactions. Conjugation of the ubiquitin-related SUMO modifier to target proteins provides a platform for protein-protein interactions and ordered assembly of multi-protein complexes. In humans, three SUMO forms (SUMO1, SUMO2, and SUMO3) can be attached to lysine residues of target proteins. Modification of proteins by SUMO are recognized by SUMO-interacting motifs termed SIMs. In natural process, polySUMOylation recruits distinct interaction partners, such as E3 ubiquitin ligases, that bind to polySUMO chains through tandem SIMs. SIMs bind to a surface patch between the α-helix and a β-sheet of the SUMO protein and extend the β-sheet of SUMO by one additional strand. the SIM either attaches as a parallel or an antiparallel strand to the SUMO β-sheet. Binding is primarily mediated by a stretch of four residues containing 3–4 hydrophobic amino acids (I, V, or L). This core interaction motif is a common property of all SIMs.
Design
Some membrane-less organelles, such as stress granules and P bodies, have been discovered in recent years. Proteins condense into droplets and assemble these organelles through a process called phase separation.