Difference between revisions of "Part:BBa J70004"
| Line 2: | Line 2: | ||
<partinfo>BBa_J70004 short</partinfo> | <partinfo>BBa_J70004 short</partinfo> | ||
| − | Cloning plasmid used by BioBasic for iGEM parts. This plasmid does not have a standard BioBrick cloning site. | + | Cloning plasmid used by BioBasic for iGEM parts. This plasmid does not have a standard BioBrick cloning site. It is '''Ampicillin''' resistant. |
| − | + | Additional Information (from BioBasic): | |
| + | * CAP protein binding site - 615-578 (compl. strand); | ||
| + | * mRNA (LacZ) starts at nt position 531 (compl. strand); | ||
| + | * lac repressor binding site -531-511 (compl. strand). | ||
| + | |||
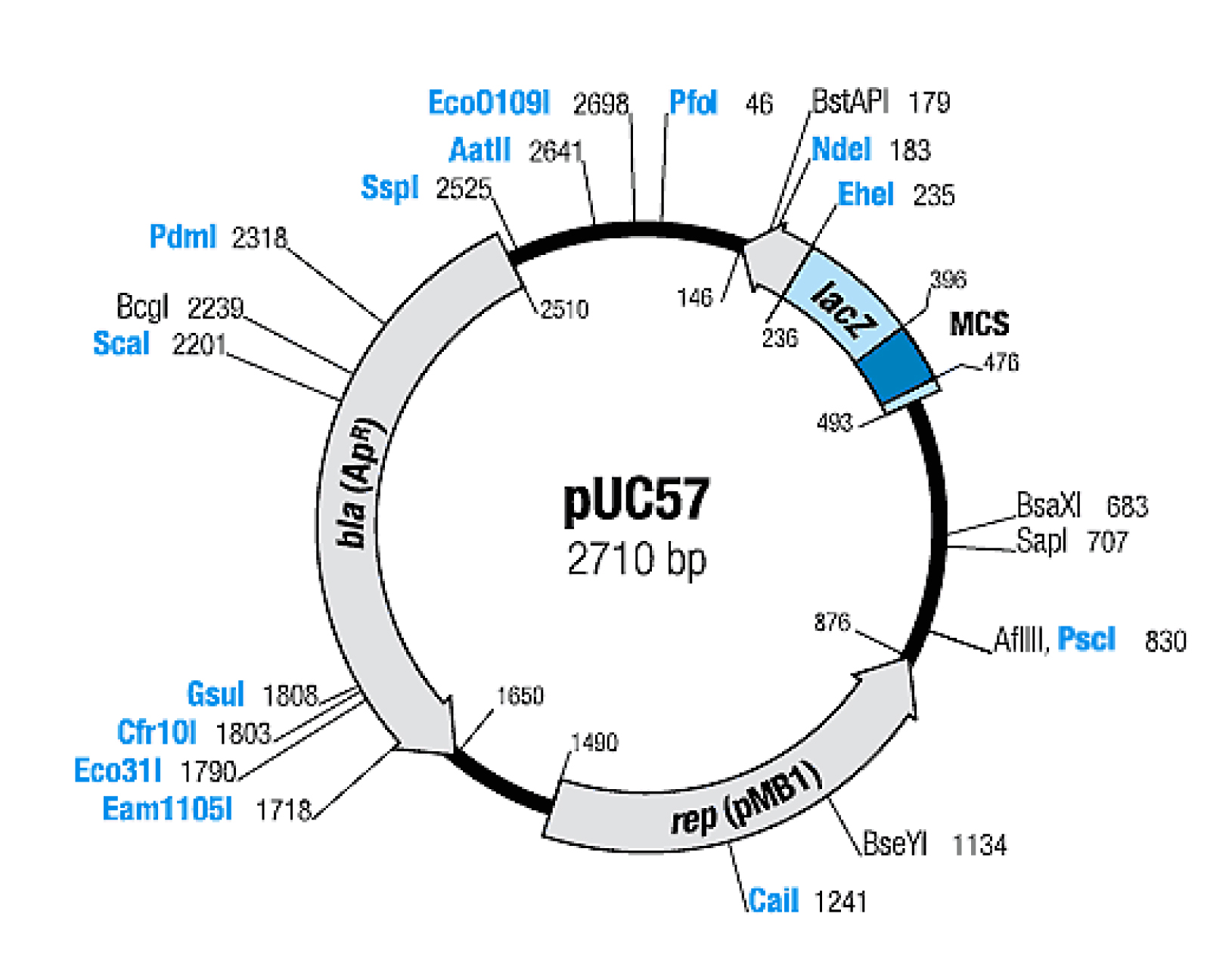
| + | Plasmid pUC57, 2710 bp in length, is a derivative of pUC19. pUC57 MCS contains 6 | ||
| + | restriction sites with protruding 3'-ends, that are resistant to E.coli exonuclease III. This | ||
| + | vector is designed for cloning and generation of ExoIII deletions. The exact position of | ||
| + | genetic elements is shown on the map (termination codons included). DNA replication | ||
| + | initiates at position 890 (+/- 1) and proceeds in indicated direction. The bla gene | ||
| + | nucleotides 2510-2442 (compl. strand) code for a single peptide. | ||
| + | |||
| + | The map (below) shows enzymes that cut pUC57 DNA once. Enzymes produced by Fermentas | ||
| + | are shown in blue. The coordinates refer to the position of the first nucleotide in each | ||
| + | recognition sequence. | ||
| + | |||
| + | [[Image:PUC57_map.jpg|300px]] | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Latest revision as of 17:41, 24 January 2008
BioBasic Cloning Plasmid pUC57
Cloning plasmid used by BioBasic for iGEM parts. This plasmid does not have a standard BioBrick cloning site. It is Ampicillin resistant.
Additional Information (from BioBasic):
- CAP protein binding site - 615-578 (compl. strand);
- mRNA (LacZ) starts at nt position 531 (compl. strand);
- lac repressor binding site -531-511 (compl. strand).
Plasmid pUC57, 2710 bp in length, is a derivative of pUC19. pUC57 MCS contains 6 restriction sites with protruding 3'-ends, that are resistant to E.coli exonuclease III. This vector is designed for cloning and generation of ExoIII deletions. The exact position of genetic elements is shown on the map (termination codons included). DNA replication initiates at position 890 (+/- 1) and proceeds in indicated direction. The bla gene nucleotides 2510-2442 (compl. strand) code for a single peptide.
The map (below) shows enzymes that cut pUC57 DNA once. Enzymes produced by Fermentas are shown in blue. The coordinates refer to the position of the first nucleotide in each recognition sequence.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal EcoRI site found at 396
Illegal XbaI site found at 425
Illegal PstI site found at 453 - 12INCOMPATIBLE WITH RFC[12]Illegal EcoRI site found at 396
Illegal PstI site found at 453 - 21INCOMPATIBLE WITH RFC[21]Illegal EcoRI site found at 396
Illegal BamHI site found at 435 - 23INCOMPATIBLE WITH RFC[23]Illegal EcoRI site found at 396
Illegal XbaI site found at 425
Illegal PstI site found at 453 - 25INCOMPATIBLE WITH RFC[25]Illegal EcoRI site found at 396
Illegal XbaI site found at 425
Illegal PstI site found at 453 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 1790
Illegal SapI site found at 707