Difference between revisions of "Part:BBa K2457002"
| Line 11: | Line 11: | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
<p>Single-guide RNA (sgRNA) is one of the two building blocks of the CRISPR-Cas9 genome editing machinery, which directs the Cas9 protein to introduce double-stranded breaks (DSB) into target genomes sequences.</p> | <p>Single-guide RNA (sgRNA) is one of the two building blocks of the CRISPR-Cas9 genome editing machinery, which directs the Cas9 protein to introduce double-stranded breaks (DSB) into target genomes sequences.</p> | ||
| − | <p>In nature, after the memory development fase made by Cas proteins, the CRISPR locus is transcribed by RNA polymerase synthesizing the crRNA precursor.The post-transcriptional maturation process begins, | + | <p>In nature, after the memory development fase made by Cas proteins, the CRISPR locus is transcribed by RNA polymerase synthesizing the crRNA precursor.The post-transcriptional maturation process begins, when this pre-crRNA ,through the complementarity its palindromic repetition, hybridizes itself with a transactivator RNA (tracrRNA), catalyzing the double strand ribonuclease formation RNAses III with the Cas9 protein. This complex cleaves the upstream palindromic repeated hairpin, making small hybridized crRNAs with the transactivator RNA (tracrRNA) in a duo composed by crRNA-tracrRNA, carrying a specific protospacer (complementary sequence) from the invasor DNA. This system, demonstrated on Figure 2 below, Deltcheva et al suggested in 2011, having as inspiration the microRNAs by interference RNAs in eukaryotes.</p> |
| − | <p>Therefore, the complex crRNA | + | <p>Therefore, the complex crRNA-tracrRNA consists of: a crRNA, with 42 nucleotides, deriving from the pre-crRNA transcribed on locus CRISPR; and a tracrRNA with 89 nucleotides, transcribed upstream to Cas operon. This non-coding RNAs hybridize themselves by Rosalind-Watson-Crick base pairing, composing a more negatively charged structure. Through polar interaction and bases stacking, the crRNA-tracrRNA is adjusted in a positive charged site between REC and NUC lobuses interface originated at Cas9 protein stemming from Streptococcus pyogenes, changing its quaternary structure to a catalytically active ribonucleoprotein complex (RNP). It has the ability to cleave DNA target sequences that are complementary to the 20 guide nucleotides present in crRNA through the HNH and RuvC nuclease domains.</p> |
===Design=== | ===Design=== | ||
<b>Synthetic sgRNA structures</b> | <b>Synthetic sgRNA structures</b> | ||
| − | <p>Nowadays, the first synthetic | + | <p>Nowadays, the first synthetic chimera of native guide RNA , built by Jinek et al in 2012, is the RNA structure commonly applied in CRISPR Cas9-based scientific projects. This structure has a shorter duplex when compared with native (Fig. 4) composed by: I) a guide sequence with 20 bp + linker loop GAAA + 30 bp of RNA-transactivator tracrRNA.</p> |
| − | <p>However, | + | <p>However, although this structure widely used containing 30 bp of tracrRNA tail is the minimum region for Cas9 interaction, in order to form its riboonucleoproteic complex catalytically active (RNP) in vitro. Guide RNAs, as an extended tail (+67 till +87), drastically enhance Cas9 activity in vivo (Dang et al. 2015)</p> |
===Characterization=== | ===Characterization=== | ||
Revision as of 21:21, 1 November 2017
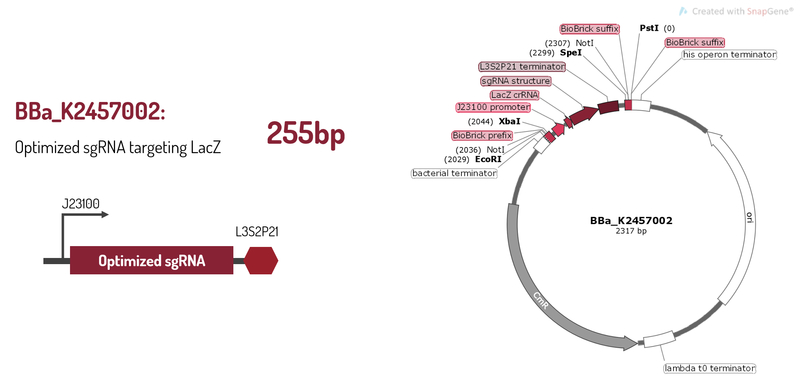
Optimized sgRNA targeting LacZ
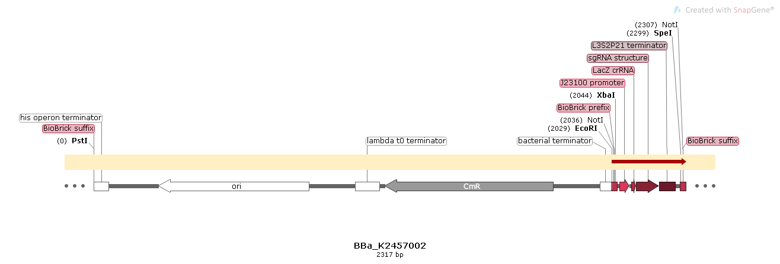
BBa_K2457002 was engineered to be a building block of the standard BioBrick toolbox for bacterial genome: the ([http://2017.igem.org/Team:Amazonas_Brazil pCRISPeasy]). It’s the first BioBrick available on Registry with an improved sgRNA sequence - both crRNA design and tracrRNA secondary structure - to increase the CRISPR-Cas9 editing efficiency. BBa_K2457002 is composed by: the J23100 constitutive promoter sequence + an optimized sgRNA (crRNA with a GG motif at the 3’ end of it’s target-specific sequence from LacZ [1] and tracrRNA with extended duplex length and a mutation at the thymine 4 [2]) + L3S2P21 terminator [3]. It was rationally designed to be standardized AND interchangeable, aiming to provide a BioBrick to attend multiple genome editing purposes. For that matter, It’s easy to engineer new modules aiming to insert information into the genome through RFC10 Assembly.
Figure 1: BBa_K2457002 circuit.
Usage and Biology
Single-guide RNA (sgRNA) is one of the two building blocks of the CRISPR-Cas9 genome editing machinery, which directs the Cas9 protein to introduce double-stranded breaks (DSB) into target genomes sequences.
In nature, after the memory development fase made by Cas proteins, the CRISPR locus is transcribed by RNA polymerase synthesizing the crRNA precursor.The post-transcriptional maturation process begins, when this pre-crRNA ,through the complementarity its palindromic repetition, hybridizes itself with a transactivator RNA (tracrRNA), catalyzing the double strand ribonuclease formation RNAses III with the Cas9 protein. This complex cleaves the upstream palindromic repeated hairpin, making small hybridized crRNAs with the transactivator RNA (tracrRNA) in a duo composed by crRNA-tracrRNA, carrying a specific protospacer (complementary sequence) from the invasor DNA. This system, demonstrated on Figure 2 below, Deltcheva et al suggested in 2011, having as inspiration the microRNAs by interference RNAs in eukaryotes.
Therefore, the complex crRNA-tracrRNA consists of: a crRNA, with 42 nucleotides, deriving from the pre-crRNA transcribed on locus CRISPR; and a tracrRNA with 89 nucleotides, transcribed upstream to Cas operon. This non-coding RNAs hybridize themselves by Rosalind-Watson-Crick base pairing, composing a more negatively charged structure. Through polar interaction and bases stacking, the crRNA-tracrRNA is adjusted in a positive charged site between REC and NUC lobuses interface originated at Cas9 protein stemming from Streptococcus pyogenes, changing its quaternary structure to a catalytically active ribonucleoprotein complex (RNP). It has the ability to cleave DNA target sequences that are complementary to the 20 guide nucleotides present in crRNA through the HNH and RuvC nuclease domains.
Design
Synthetic sgRNA structures
Nowadays, the first synthetic chimera of native guide RNA , built by Jinek et al in 2012, is the RNA structure commonly applied in CRISPR Cas9-based scientific projects. This structure has a shorter duplex when compared with native (Fig. 4) composed by: I) a guide sequence with 20 bp + linker loop GAAA + 30 bp of RNA-transactivator tracrRNA.
However, although this structure widely used containing 30 bp of tracrRNA tail is the minimum region for Cas9 interaction, in order to form its riboonucleoproteic complex catalytically active (RNP) in vitro. Guide RNAs, as an extended tail (+67 till +87), drastically enhance Cas9 activity in vivo (Dang et al. 2015)
Characterization
Sequencing
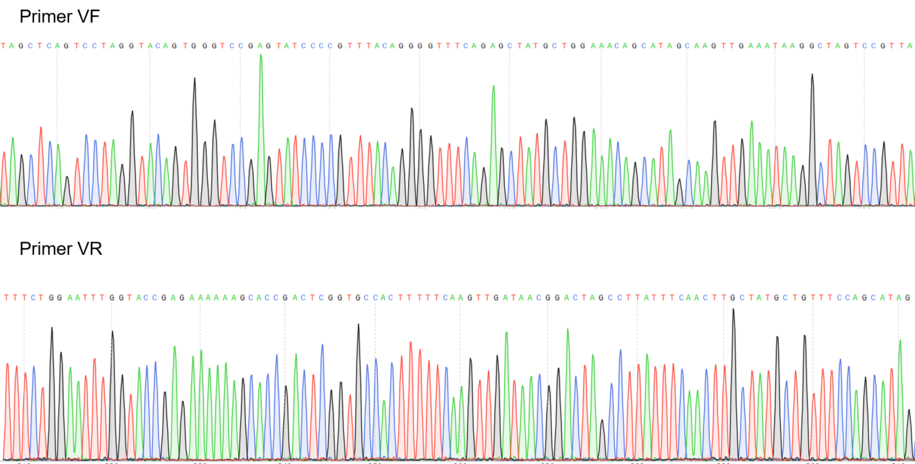 Figure 2: Sequencing electropherogram from BBa_K2457002.
Figure 2: Sequencing electropherogram from BBa_K2457002.
Figure 3: Alignment of the designed sequence and our final construction from BBa_K2457002.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]