Difference between revisions of "Part:BBa K2201373"
| Line 38: | Line 38: | ||
The second system is our genetic circuit. In this system the CDS of the target gene is in front of a CDS for a T3 DNA-polymerase. Thus the expression of the target gene is nearly on the same level as the expression of the target gene. The expressed T3 RNA-polymerase expresses the mRFP under the control of the T3-promoter. | The second system is our genetic circuit. In this system the CDS of the target gene is in front of a CDS for a T3 DNA-polymerase. Thus the expression of the target gene is nearly on the same level as the expression of the target gene. The expressed T3 RNA-polymerase expresses the mRFP under the control of the T3-promoter. | ||
To demonstrate the advantages of our new construct we modelled the amount of mRFP-transcript for both constructs. | To demonstrate the advantages of our new construct we modelled the amount of mRFP-transcript for both constructs. | ||
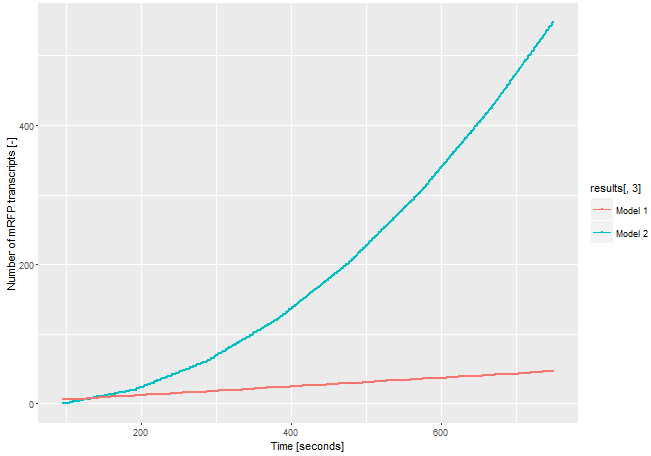
| − | If we assume the expression of the gene of interest is low and only one <i>E. coli</i> RNA-polymerase with a chain elongation rate of 50 nucleotides per second translates the both products, construct 1 produces 1 mRFP-transcript in 16 seconds. Construct 2 expresses 1 T3 RNA-polymerase-transcript every 52 seconds. After translation (with a translation rate of 20 amino acids per second ~42 sec), these polymerases transcribe mRFP-transcript with a chain elongation rate of 170 nucleotides per second. So every T3 RNA-polymerase expresses one mRFP-transcript every 4.7 seconds. The resulting amount of mRFP transcripts is shown in Figure | + | If we assume the expression of the gene of interest is low and only one <i>E. coli</i> RNA-polymerase with a chain elongation rate of 50 nucleotides per second translates the both products, construct 1 produces 1 mRFP-transcript in 16 seconds. Construct 2 expresses 1 T3 RNA-polymerase-transcript every 52 seconds. After translation (with a translation rate of 20 amino acids per second ~42 sec), these polymerases transcribe mRFP-transcript with a chain elongation rate of 170 nucleotides per second. So every T3 RNA-polymerase expresses one mRFP-transcript every 4.7 seconds. The resulting amount of mRFP transcripts is shown in Figure 3. The script for our modelling can be found <html><a href="https://static.igem.org/mediawiki/parts/8/8a/T--Bielefeld-CeBiTec--model-composite-2.txt">here</a></html>. |
| − | [[File:T--Bielefeld-CeBiTec--model-composite.png|600px|thumb|center|<b>Figure | + | [[File:T--Bielefeld-CeBiTec--model-composite.png|600px|thumb|center|<b>Figure 3:</b> Modelling on the amount of mRFP-transcript transcribed through the two different models.]]<br> |
The number of mRFP transcripts is a lot higher with construct 2 and this model leaves out the fact that one transcript of the T3 RNA-polymerase transcript could be translated severel times and enhance the signal even more. Despite this model is simplified, it shows the advantages of using the T3 RNA-polymerase for signal enhancing. | The number of mRFP transcripts is a lot higher with construct 2 and this model leaves out the fact that one transcript of the T3 RNA-polymerase transcript could be translated severel times and enhance the signal even more. Despite this model is simplified, it shows the advantages of using the T3 RNA-polymerase for signal enhancing. | ||
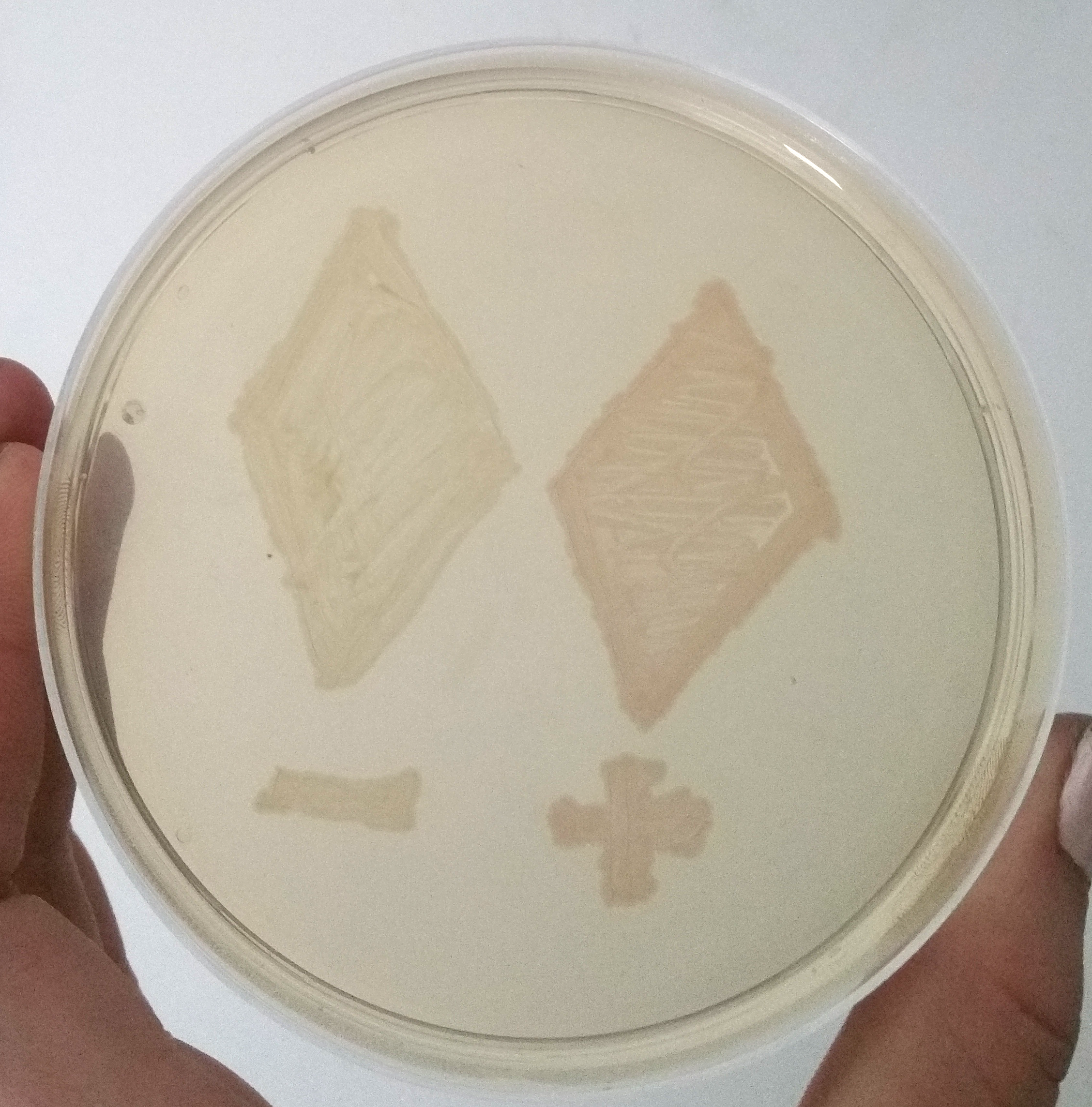
| − | [[File:T--Bielefeld-CeBiTec--composite-1.png|600px|thumb|center|<b>Figure | + | [[File:T--Bielefeld-CeBiTec--composite-1.png|600px|thumb|center|<b>Figure 4:</b> Two Smear of two clones containing only mRFP under an uninduced T7 promoter and containing the mRFP enhancing system under the same promotor, after 12 h of incubation at 37 °C.]]<br> |
| − | To demonstrate the signal enahncing system we cloned the part E1010 and our signal enhancing system (BBa_K2201373) behind the part BBa_K2201900 containing our positive selection plasmid. A smear of the transformands is shown in Figure | + | To demonstrate the signal enahncing system we cloned the part E1010 and our signal enhancing system (BBa_K2201373) behind the part BBa_K2201900 containing our positive selection plasmid. A smear of the transformands is shown in Figure 4. The transcription of the promotor is only a basal transcription, thus, it is a weak transcription. Despite this weak transcription, the expressed mRFP of the signal enhancing system is clearly visible, while the fluorescence of only the mRFP is not visible. |
| − | [[File:T--Bielefeld-CeBiTec--composite-2.png|600px|thumb|center|<b>Figure | + | [[File:T--Bielefeld-CeBiTec--composite-2.png|600px|thumb|center|<b>Figure 5:</b> Picture of a negative selection round. The clon still containing the positive selection plasmid, thus the mRFP enhancing system, is red.]]<br> |
For the selection of tRNA/aminoacylsynthetase for the incorporation of non-canonical amino acids we needed to check if cells still contain the positive selection plasmid in the negative selection round. Therefore, we decided to incorporate the mRFP signal enhancing system behind the CDS of our positive selection plasmid BBa_K2201900. If the cells contain this plasmid in the negative selection, they should be visible as red clones. In contrast, the clones containing the negative selection plasmid should be coulourless. A picture of one round of the negative selection, shown in Figure 5, shows that one cell is red. Thus this cell contains the positive selection plasmid. | For the selection of tRNA/aminoacylsynthetase for the incorporation of non-canonical amino acids we needed to check if cells still contain the positive selection plasmid in the negative selection round. Therefore, we decided to incorporate the mRFP signal enhancing system behind the CDS of our positive selection plasmid BBa_K2201900. If the cells contain this plasmid in the negative selection, they should be visible as red clones. In contrast, the clones containing the negative selection plasmid should be coulourless. A picture of one round of the negative selection, shown in Figure 5, shows that one cell is red. Thus this cell contains the positive selection plasmid. | ||
Revision as of 14:22, 31 October 2017
T3 polymerase with inverted mRFP under T3 promoter control for signal enhancing
This part contains a T3 RNA-Polymerase with an reversed mRFP under T3 RNA-polymerase control for signal enhancing. It is an improved reporter and a genetic circuit that could report even weak expression levels. This part was created following the model of an amplifier in electrical engineering to intensify an input signal and could be used in a broad range of synthetic biology applications. We used this part for our selection system for the incorporation of non-canonical amino acids
Usage and Biology
The T3 RNA-polymerase is highly specific to T3 promoters and orthogonal to the T7 RNA-polymerase. Thus it can be widely utilized in synthetic biology, even in expression strains which encode for a T7 RNA-polymerase like E. coli BL21. We wanted to use this polymerase to create a reporter for the use in genetic circuits to make even weak expression visible.
At the moment fluorescent proteins with an emission wavelength within the visible spectra are used to report expression of the gene of interest. Therefor the CDS of the fluorescent protein is placed behind the CDS of the target protein without a terminator or promoter in between. Due to that, the expression level of the target protein is nearly the same as the expression of the fluorescent protein. The fluorescence of the protein could be used to find out if the gene of interest is translated. This works fine when the expression of the target protein is strong enough to build enough fluorescent protein to generate a strong fluorescent signal.
In a lot of applications only a weak expression is, or should be, reached. For our project, we needed a reliable reporter to detect the expression of our gene of interest on our selection plasmid. For our selection system it is important that the genes are only expressed on a weak level. If the CDS of mRFP is placed behind the CDS of this gene, no fluorescence was visible. Through the function of the gene of interest we knew it was expressed. To solve this reporter problem we intended to build a genetic circuit following the model of an amplifier used in electrical engineering.
Easy amplifiers have already been built by iGEM Cambridge 2009. They build a simple circuit using an activator which leads to the transcription of a reporter gene on a higher level as under the first promoter. To explain their system they used the term Polymerases per second (PoPs). PoPS is the flow of RNA polymerase molecules along DNA (i.e., 'current' for gene expression). The PoPS level is set by the amount of RNA polymerase molecules that pass a specific position on DNA each second. The system is shown in Figure 1 and should enhance the level of input PoPs.

Figure 1: Signal strenthening system of iGEM Cambridge 2009.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 2717
Illegal AgeI site found at 2829 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 2585
Functional Parameters
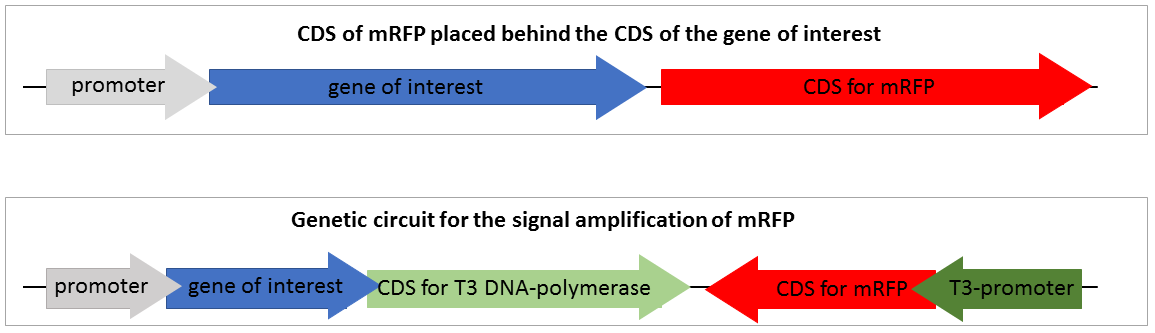
To design a genetic circuit that amplifiers a reporter signal we decided to use an RNA-polymerase, orthogonal to the native E.coli RNA-polymerase. We used the T3 RNA-polymerase which characterised by iGEM Peking 2010. The T3 RNA-polymerase is highly specific to T3-promoters and orthogonal to the E. coli DNA-polymerases and even to the T7 RNA-polymerase which is very similar. We decided not to use the T7 RNA-polymerase because of it is often used for recombinant expression and we wanted to provide a system which could be used in expression strains like BL21. The construction of our designed composite part and the standard mRFP reporter are shown in Figure 2.
Figure 2 shows two constructions of reporters for gene expression. The first one is the standard reporter for the gene expression of the target gene, using mRFP as reporter. If the gene of interest is expressed, the mRFP is expressed on nearly the same level. Through the detection of the mRFP fluorescence the gene expression of the gene of interest could be detected. This system is suitable for high expression rates, but if the expression of the target gene is only on a weak expression level, the fluorescence of the expressed mRFP is too low to detect and thus not visible.
The second system is our genetic circuit. In this system the CDS of the target gene is in front of a CDS for a T3 DNA-polymerase. Thus the expression of the target gene is nearly on the same level as the expression of the target gene. The expressed T3 RNA-polymerase expresses the mRFP under the control of the T3-promoter. To demonstrate the advantages of our new construct we modelled the amount of mRFP-transcript for both constructs. If we assume the expression of the gene of interest is low and only one E. coli RNA-polymerase with a chain elongation rate of 50 nucleotides per second translates the both products, construct 1 produces 1 mRFP-transcript in 16 seconds. Construct 2 expresses 1 T3 RNA-polymerase-transcript every 52 seconds. After translation (with a translation rate of 20 amino acids per second ~42 sec), these polymerases transcribe mRFP-transcript with a chain elongation rate of 170 nucleotides per second. So every T3 RNA-polymerase expresses one mRFP-transcript every 4.7 seconds. The resulting amount of mRFP transcripts is shown in Figure 3. The script for our modelling can be found here.
The number of mRFP transcripts is a lot higher with construct 2 and this model leaves out the fact that one transcript of the T3 RNA-polymerase transcript could be translated severel times and enhance the signal even more. Despite this model is simplified, it shows the advantages of using the T3 RNA-polymerase for signal enhancing.
To demonstrate the signal enahncing system we cloned the part E1010 and our signal enhancing system (BBa_K2201373) behind the part BBa_K2201900 containing our positive selection plasmid. A smear of the transformands is shown in Figure 4. The transcription of the promotor is only a basal transcription, thus, it is a weak transcription. Despite this weak transcription, the expressed mRFP of the signal enhancing system is clearly visible, while the fluorescence of only the mRFP is not visible.
For the selection of tRNA/aminoacylsynthetase for the incorporation of non-canonical amino acids we needed to check if cells still contain the positive selection plasmid in the negative selection round. Therefore, we decided to incorporate the mRFP signal enhancing system behind the CDS of our positive selection plasmid BBa_K2201900. If the cells contain this plasmid in the negative selection, they should be visible as red clones. In contrast, the clones containing the negative selection plasmid should be coulourless. A picture of one round of the negative selection, shown in Figure 5, shows that one cell is red. Thus this cell contains the positive selection plasmid.
Apart from our application for this part, there are a lot of applications suitable for this signal enhancing part. Despite all applications where even weak expression should be reported, it is a usefull method which could be transfered to a lot of problems in synthetic biology requiering a stronger output signal.