Difference between revisions of "Part:BBa K2201004"
| Line 40: | Line 40: | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| − | + | <html> | |
| + | <p align="justify"> | ||
Phaeodactylum tricornutum, a diatom of the genus Phaedactylum, features six putative nucleotide transporters (NTTs). Two isoforms of these NTTs have been characterized by Ast <i>et al.</i> 2009 and it was shown that both isoforms facilitate transport across the plastid membrane. While isoform 1 (NTT1) acts as a proton-dependent adenine nucleotide importer, NTT2 facilitates the counter exchange of (deoxy-)nucleoside triphosphates (Ast <i>et al.</i>, 2009).The isoform 2 of the nucleotide transporter was shown to be a broad range (deoxy-)nucleoside transporter, facilitating the uptake of CTP, GTP, dCTP, ATP, UTP, dGTP, dATP and TTP when expressed in <i>E. coli</i>.Zhang <i>et al.</i> 2017 investigated the use of <i>Pt</i>NTT2 for the uptake of the unnatural bases dNaM and dTPT3. Therefore, the expression of <i>Pt</i>NTT2 was investigated in different strains, under control of different promotors, and plasmid-bound as well as integrated into the chromosome. In their final design, Zhang and colleagues integrated <i>Pt</i>NTT2 chromosomally in <i>E. coli</i> BL21(DE3) under control of the lacUV5 promoter. To demonstrate its feasibility for the uptake of nucleotides in <i>E. coli</i> from the media, uptake of [α 32P]-dATP was measured. The native sequence of <i>Pt</i>NTT2 features an N-terminal signal sequence directing the subcellular localization to the plastid membrane. In <i>E. coli</i>, this signal sequence is likely to be retained, leading to a growth defect in cells expressing the native <i>Pt</i>NTT2 transporter. | Phaeodactylum tricornutum, a diatom of the genus Phaedactylum, features six putative nucleotide transporters (NTTs). Two isoforms of these NTTs have been characterized by Ast <i>et al.</i> 2009 and it was shown that both isoforms facilitate transport across the plastid membrane. While isoform 1 (NTT1) acts as a proton-dependent adenine nucleotide importer, NTT2 facilitates the counter exchange of (deoxy-)nucleoside triphosphates (Ast <i>et al.</i>, 2009).The isoform 2 of the nucleotide transporter was shown to be a broad range (deoxy-)nucleoside transporter, facilitating the uptake of CTP, GTP, dCTP, ATP, UTP, dGTP, dATP and TTP when expressed in <i>E. coli</i>.Zhang <i>et al.</i> 2017 investigated the use of <i>Pt</i>NTT2 for the uptake of the unnatural bases dNaM and dTPT3. Therefore, the expression of <i>Pt</i>NTT2 was investigated in different strains, under control of different promotors, and plasmid-bound as well as integrated into the chromosome. In their final design, Zhang and colleagues integrated <i>Pt</i>NTT2 chromosomally in <i>E. coli</i> BL21(DE3) under control of the lacUV5 promoter. To demonstrate its feasibility for the uptake of nucleotides in <i>E. coli</i> from the media, uptake of [α 32P]-dATP was measured. The native sequence of <i>Pt</i>NTT2 features an N-terminal signal sequence directing the subcellular localization to the plastid membrane. In <i>E. coli</i>, this signal sequence is likely to be retained, leading to a growth defect in cells expressing the native <i>Pt</i>NTT2 transporter. | ||
| + | </p> | ||
| + | <br style="clear: both" /> | ||
| + | </html> | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
| Line 54: | Line 58: | ||
==Computational Analysis of <i>Pt</i>NTT2== | ==Computational Analysis of <i>Pt</i>NTT2== | ||
| − | |||
| − | |||
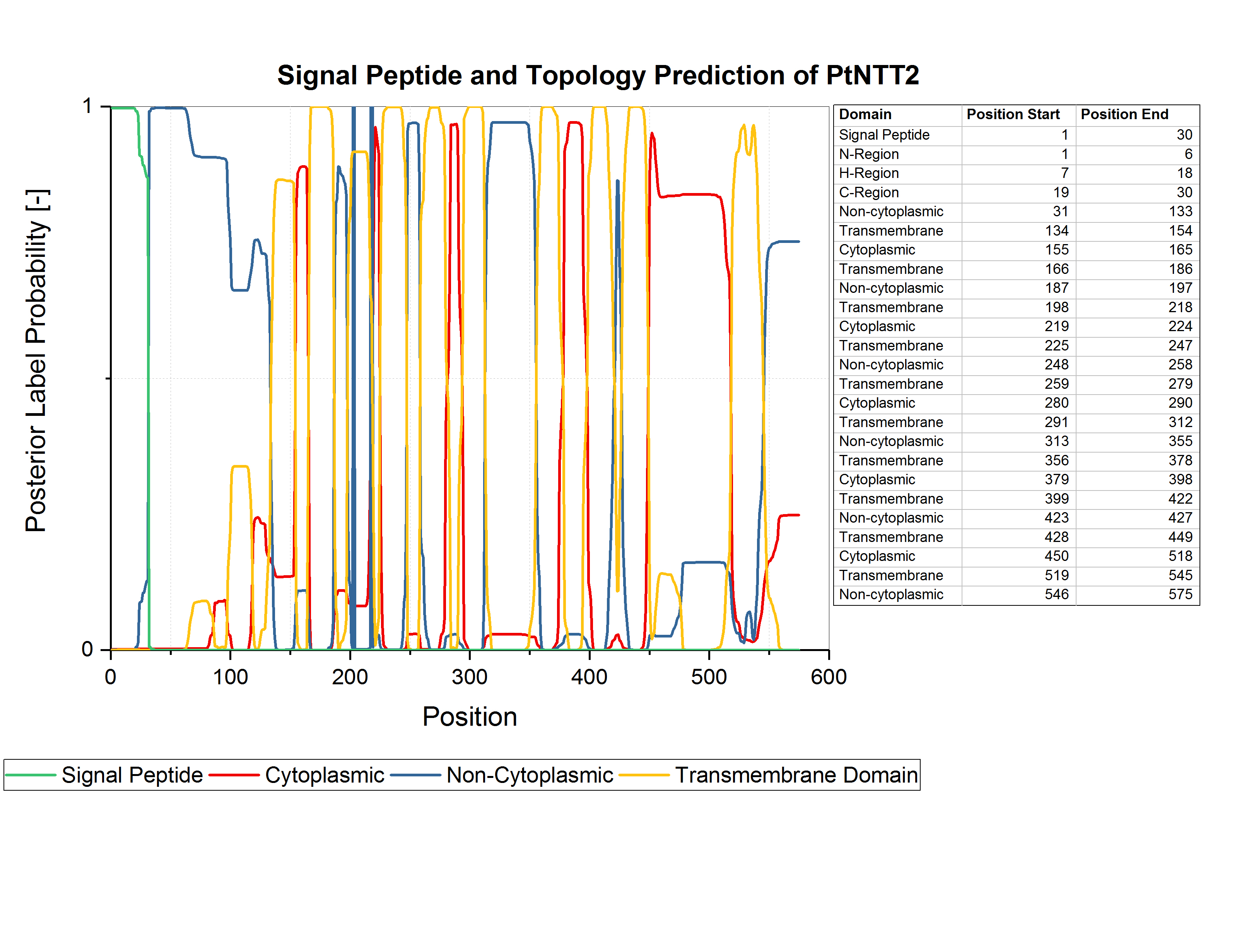
[[file:T--Bielefeld-CeBiTec--Parts PhobiusPrediction300.jpeg|600px|thumb|left|<b>Figure (1): Results of the analysis of <i>Pt</i>NTT2 using Phobius.</b><br> The 30 first amino acids are clearly recognized as a signal peptide. Ten transmembrane domains were predicted.]] | [[file:T--Bielefeld-CeBiTec--Parts PhobiusPrediction300.jpeg|600px|thumb|left|<b>Figure (1): Results of the analysis of <i>Pt</i>NTT2 using Phobius.</b><br> The 30 first amino acids are clearly recognized as a signal peptide. Ten transmembrane domains were predicted.]] | ||
| − | + | <html> | |
| + | <p align="justify"> | ||
Zhang <i>et al.</i> used an N-terminal truncated version of <i>Pt</i>NTT2, lacking the first 65 amino acids, since they observed some kind of toxicity resulting from the native N-terminal sequence (Zhang et al., 2017). For our project, we analyzed the amino acid sequence of <i>Pt</i>NTT2 using the prediction software Phobius (Käll et al., 2007). Using Phobius, we analyzed the signal peptide and the transmembrane topology of <i>Pt</i>NTT2. The analysis revealed that the native signal peptide is formed by amino acids 1 30, which means that Zhang et al. removed more than the native signal peptide for their experiment. The results of the prediction are shown in figure (1). Analysis of the transmembrane topology of the transporter, which is integrated into the plastid membrane in its native algal cell, shows iterative non-cytoplasmatic, transmembrane and cytoplasmic regions. The topology might indicate, that the transporter will be integrated into the inner membrane when expressed in <i>E. coli</i>. | Zhang <i>et al.</i> used an N-terminal truncated version of <i>Pt</i>NTT2, lacking the first 65 amino acids, since they observed some kind of toxicity resulting from the native N-terminal sequence (Zhang et al., 2017). For our project, we analyzed the amino acid sequence of <i>Pt</i>NTT2 using the prediction software Phobius (Käll et al., 2007). Using Phobius, we analyzed the signal peptide and the transmembrane topology of <i>Pt</i>NTT2. The analysis revealed that the native signal peptide is formed by amino acids 1 30, which means that Zhang et al. removed more than the native signal peptide for their experiment. The results of the prediction are shown in figure (1). Analysis of the transmembrane topology of the transporter, which is integrated into the plastid membrane in its native algal cell, shows iterative non-cytoplasmatic, transmembrane and cytoplasmic regions. The topology might indicate, that the transporter will be integrated into the inner membrane when expressed in <i>E. coli</i>. | ||
</p> | </p> | ||
<br style="clear: both" /> | <br style="clear: both" /> | ||
| + | </html> | ||
==Plasmid Design== | ==Plasmid Design== | ||
| + | <html> | ||
| + | <p align="justify"> | ||
For the analysis and characterization of <i>Pt</i>NTT2, a total of eleven plasmids were designed and cloned based on initial research and the computational analysis. The coding sequence of <i>Pt</i>NTT2 was codon optimized using the IDT Codon Optimization Tool and ordered as two gBlocks. Using overlap extension PCR, the two gBlocks were put together and inserted into pSB1C3 using Gibson Assembly. The truncated versions of the transporter as well as the versions with new signal peptides were constructed using primers and Gibson Assembly. | For the analysis and characterization of <i>Pt</i>NTT2, a total of eleven plasmids were designed and cloned based on initial research and the computational analysis. The coding sequence of <i>Pt</i>NTT2 was codon optimized using the IDT Codon Optimization Tool and ordered as two gBlocks. Using overlap extension PCR, the two gBlocks were put together and inserted into pSB1C3 using Gibson Assembly. The truncated versions of the transporter as well as the versions with new signal peptides were constructed using primers and Gibson Assembly. | ||
| + | </p> | ||
| + | </html> | ||
<html> | <html> | ||
| Line 108: | Line 116: | ||
<td class="td">pSB1C3-PlacUV5-PtNTT2-GFP</td> | <td class="td">pSB1C3-PlacUV5-PtNTT2-GFP</td> | ||
<td class="td"><a href="https://parts.igem.org/Part:BBa_K2201002">BBa_K2201002</a></td> | <td class="td"><a href="https://parts.igem.org/Part:BBa_K2201002">BBa_K2201002</a></td> | ||
| − | <td class="td">Fusion protein of BBa_ K2201000 with GFP (<a href="https://parts.igem.org/Part:BBa_E0040"> BBa_E0040 </a>), | + | <td class="td">Fusion protein of BBa_ K2201000 with GFP (<a href="https://parts.igem.org/Part:BBa_E0040"> BBa_E0040 </a>), cMyc epitope tag as linker (<a href="https://parts.igem.org/Part:BBa_K2201181"> BBa_K2201181 </a>)</td> |
</tr> | </tr> | ||
<tr class="tr"> | <tr class="tr"> | ||
<td class="td">pSB1C3-PlacUV5-PtNTT2(66-575)-GFP</td> | <td class="td">pSB1C3-PlacUV5-PtNTT2(66-575)-GFP</td> | ||
<td class="td"><a href="https://parts.igem.org/Part:BBa_K2201003">BBa_K2201003</a></td> | <td class="td"><a href="https://parts.igem.org/Part:BBa_K2201003">BBa_K2201003</a></td> | ||
| − | <td class="td">Fusion protein of BBa_ K2201001 with GFP (<a href="https://parts.igem.org/Part:BBa_E0040"> BBa_E0040 </a>), | + | <td class="td">Fusion protein of BBa_ K2201001 with GFP (<a href="https://parts.igem.org/Part:BBa_E0040"> BBa_E0040 </a>), cMyc epitope tag as linker (<a href="https://parts.igem.org/Part:BBa_K2201181"> BBa_K2201181 </a>)</td> |
</tr> | </tr> | ||
<tr class="tr"> | <tr class="tr"> | ||
<td class="td">pSB1C3-PlacUV5-PtNTT2(31-575)-GFP</td> | <td class="td">pSB1C3-PlacUV5-PtNTT2(31-575)-GFP</td> | ||
<td class="td"><a href="https://parts.igem.org/Part:BBa_K2201011">BBa_K2201011</a></td> | <td class="td"><a href="https://parts.igem.org/Part:BBa_K2201011">BBa_K2201011</a></td> | ||
| − | <td class="td">Fusion protein of BBa_K2201005 with GFP (<a href="https://parts.igem.org/Part:BBa_E0040"> BBa_E0040 </a>), | + | <td class="td">Fusion protein of BBa_K2201005 with GFP (<a href="https://parts.igem.org/Part:BBa_E0040"> BBa_E0040 </a>), cMyc epitope tag as linker (<a href="https://parts.igem.org/Part:BBa_K2201181"> BBa_K2201181 </a>)</td> |
</tr> | </tr> | ||
<tr class="tr"> | <tr class="tr"> | ||
<td class="td">pSB1C3-PlacUV5-pelB-SP-PtNTT2-GFP</td> | <td class="td">pSB1C3-PlacUV5-pelB-SP-PtNTT2-GFP</td> | ||
<td class="td"><a href="https://parts.igem.org/Part:BBa_K2201012">BBa_K2201012</a></td> | <td class="td"><a href="https://parts.igem.org/Part:BBa_K2201012">BBa_K2201012</a></td> | ||
| − | <td class="td">Fusion protein of BBa_K2201006 with GFP (<a href="https://parts.igem.org/Part:BBa_E0040"> BBa_E0040 </a>), | + | <td class="td">Fusion protein of BBa_K2201006 with GFP (<a href="https://parts.igem.org/Part:BBa_E0040"> BBa_E0040 </a>), cMyc epitope tag as linker (<a href="https://parts.igem.org/Part:BBa_K2201181"> BBa_K2201181 </a>)</td> |
</tr> | </tr> | ||
<tr class="tr"> | <tr class="tr"> | ||
<td class="td">pSB1C3-PlacUV5-TAT-SP-PtNTT2-GFP</td> | <td class="td">pSB1C3-PlacUV5-TAT-SP-PtNTT2-GFP</td> | ||
<td class="td"><a href="https://parts.igem.org/Part:BBa_K2201013">BBa_K2201013</a></td> | <td class="td"><a href="https://parts.igem.org/Part:BBa_K2201013">BBa_K2201013</a></td> | ||
| − | <td class="td">Fusion protein of BBa_K2201007 with GFP (<a href="https://parts.igem.org/Part:BBa_E0040"> BBa_E0040 </a>), | + | <td class="td">Fusion protein of BBa_K2201007 with GFP (<a href="https://parts.igem.org/Part:BBa_E0040"> BBa_E0040 </a>), cMyc epitope tag as linker (<a href="https://parts.igem.org/Part:BBa_K2201181"> BBa_K2201181 </a>)</td> |
</tr> | </tr> | ||
</tbody> | </tbody> | ||
| Line 138: | Line 146: | ||
===Shake Flask Cultivation=== | ===Shake Flask Cultivation=== | ||
| + | [[File:T--Bielefeld-CeBiTec--Parts Shakeflask.jpeg|500px|thumb|right|<b>Figure (3): Shake flask cultivation of all <i>Pt</i>NTT2 variants. </b><br> <i>E. coli</i> BL21(DE3) and <i>E. coli</i> BL21(DE3) pSB1C3-PtNTT2, not expressing <i>Pt</i>NTT2, were used as negative controls. Two biological replicates of each strain were cultivated and three technical replicates taken for each measurement. A clear difference in the growth rates can be observed, with <i>E. coli</i> BL21(DE3) pSB1C3-PlacUV5-PtNTT2 and <i>E. coli</i> BL21(DE3) pSB1C3-PlacUV5-TAT-SP-PtNTT2 showing the weakest growth. Both strains also show the longest lag phase, which is nearly twice as long as the lag phase of <i>E. coli</i> BL21(DE3). <i>E. coli</i> BL21(DE3) pSB1C3-PlacUV5-PtNTT2(66-575) and <i>E. coli</i> BL21(DE3) pSB1C3-PlacUV5-pelB-SP-PtNTT2 show the best growth of all <i>Pt</i>NTT2 variants, reaching the highest OD<sub>600</sub>.]] | ||
| + | <html> | ||
| + | <p align="justify"> | ||
| + | |||
| + | Given that Zhang <i>et al.</i>, 2017 reported some kind of toxicity resulting from the N-terminal sequence of <i>Pt</i>NTT2, we first investigated if we could also observe the same toxicity associated with the N-terminal sequence. We also tested whether our own versions of the transporter result in better growth and reduced toxicity compared to the native transporter. Therefore, after cloning the plasmids in <i>E. coli</i> DH5α and verifying the correct assembly via sequencing, all plasmids were transformed into <i>E. coli</i> BL21(DE3). The presence of the correct plasmids was again verified by colony PCR. | ||
| + | The first cultivations were carried out in shake flasks in LB media. A total cultivation volume of 50 mL was used. The cultures were incubated at 37 °C and 180 rpm. All cultures were inoculated with an OD<sub>600</sub> of 0.01. OD<sub>600</sub> was measured every hour during lag and stationary phase and every 30 minutes during the exponential phase. The optical density was measured using an Eppendorf Photometer and standard cuvettes. <i>E. coli</i> BL21(DE3) without a plasmid and <i>E. coli</i> BL21(DE3) harboring pSB1C3-PtNTT2 were used as negative controls. Two biological replicates of each strain were tested and three technical replicates were measured for each timepoint. | ||
| + | |||
| + | <i>E. coli</i> BL21(DE3) without any plasmid shows the best growth, with the highest specific growth rate and the highest final OD<sub>600</sub> of 5.178 ± 0.046. The second negative control, <i>E. coli</i> BL21(DE3) harboring pSB1C3-PtNTT2 (BBa_K2201004) reached the second highest OD<sub>600</sub> with 4.638 ± 0.029. Of the functional <i>Pt</i>NTT2 variants, strains harboring pSB1C3-PlacUV5-PtNTT2(66-575) (BBa_K2201001) and pSB1C3-PlacUV5-pelB-SP-PtNTT2 (BBa_K2201006) reached the highest ODs with 4.397 ± 0.062 and 4.171 ± 0.051, respectively. <i>E. coli</i> BL21(DE3) pSB1C3-PlacUV5-PtNTT2(31-575) (BBa_K2201005) showed similar growth to the two previous strains during the lag phase and early exponential phase, but reaching a lower OD<sub>600</sub> of 3.802 ± 0.135. <i>E. coli</i> BL21(DE3) pSB1C3-PlacUV5-PtNTT2 (BBa_K2201000) and <i>E. coli</i> BL21(DE3) pSB1C3-PlacUV5-TAT-SP-PtNTT2 (BBa_K2201007) grew significantly weaker compared to all previous strains, with a lag phase nearly four hours long and final ODs of 2.499 ± 0.134 and 2.735 ± 0.150, respectively. The difference in growth between the two negative controls can be explained by the metabolic burden caused by plasmid replication and expression of the chloramphenicol resistance. Therefore, <i>E. coli</i> BL21(DE3) pSB1C3-PtNTT2 is the more accurate control, since all samples contain the same plasmid backbone and were also grown in LB media supplemented with chloramphenicol. The difference in growth between strains expressing the native, full length transporter and the truncated version <i>Pt</i>NTT2(66-575), observed by Zhang et al. 2017, could also be shown. This negative effect might be associated with the native signal peptide of <i>Pt</i>NTT2, which <i>E. coli</i> might not be able to process correctly. Another explanation for the weak growth could be that the native transporter variant has a higher activity compared to the other variants. If the activity is too high, this might lead to a toxic effect and to the observed weak growth. | ||
| + | |||
| + | |||
| + | </p> | ||
| + | <br style="clear: both" /> | ||
| + | </html> | ||
===Microcultivations=== | ===Microcultivations=== | ||
Revision as of 14:55, 30 October 2017
Nucleotide Transporter PtNTT2 from Phaeodactylum tricornutum
Usage and Biology
Phaeodactylum tricornutum, a diatom of the genus Phaedactylum, features six putative nucleotide transporters (NTTs). Two isoforms of these NTTs have been characterized by Ast et al. 2009 and it was shown that both isoforms facilitate transport across the plastid membrane. While isoform 1 (NTT1) acts as a proton-dependent adenine nucleotide importer, NTT2 facilitates the counter exchange of (deoxy-)nucleoside triphosphates (Ast et al., 2009).The isoform 2 of the nucleotide transporter was shown to be a broad range (deoxy-)nucleoside transporter, facilitating the uptake of CTP, GTP, dCTP, ATP, UTP, dGTP, dATP and TTP when expressed in E. coli.Zhang et al. 2017 investigated the use of PtNTT2 for the uptake of the unnatural bases dNaM and dTPT3. Therefore, the expression of PtNTT2 was investigated in different strains, under control of different promotors, and plasmid-bound as well as integrated into the chromosome. In their final design, Zhang and colleagues integrated PtNTT2 chromosomally in E. coli BL21(DE3) under control of the lacUV5 promoter. To demonstrate its feasibility for the uptake of nucleotides in E. coli from the media, uptake of [α 32P]-dATP was measured. The native sequence of PtNTT2 features an N-terminal signal sequence directing the subcellular localization to the plastid membrane. In E. coli, this signal sequence is likely to be retained, leading to a growth defect in cells expressing the native PtNTT2 transporter.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Contents
Computational Analysis of PtNTT2
Zhang et al. used an N-terminal truncated version of PtNTT2, lacking the first 65 amino acids, since they observed some kind of toxicity resulting from the native N-terminal sequence (Zhang et al., 2017). For our project, we analyzed the amino acid sequence of PtNTT2 using the prediction software Phobius (Käll et al., 2007). Using Phobius, we analyzed the signal peptide and the transmembrane topology of PtNTT2. The analysis revealed that the native signal peptide is formed by amino acids 1 30, which means that Zhang et al. removed more than the native signal peptide for their experiment. The results of the prediction are shown in figure (1). Analysis of the transmembrane topology of the transporter, which is integrated into the plastid membrane in its native algal cell, shows iterative non-cytoplasmatic, transmembrane and cytoplasmic regions. The topology might indicate, that the transporter will be integrated into the inner membrane when expressed in E. coli.
Plasmid Design
For the analysis and characterization of PtNTT2, a total of eleven plasmids were designed and cloned based on initial research and the computational analysis. The coding sequence of PtNTT2 was codon optimized using the IDT Codon Optimization Tool and ordered as two gBlocks. Using overlap extension PCR, the two gBlocks were put together and inserted into pSB1C3 using Gibson Assembly. The truncated versions of the transporter as well as the versions with new signal peptides were constructed using primers and Gibson Assembly.
| Plasmid Name | BioBrick Number | Characteristics |
|---|---|---|
| pSB1C3-PtNTT2 | BBa_K2201004 | Only the cds |
| pSB1C3-PlacUV5-PtNTT2 | BBa_K2201000 | cds with lacUV5 promotor and a strong RBS ( BBa_B0034 ) |
| pSB1C3-PlacUV5-PtNTT2(66-575) | BBa_K2201001 | cds with lacUV5 promotor and a strong RBS ( BBa_B0034 ),truncated version lacking the first 65 amino acids |
| pSB1C3-PlacUV5-PtNTT2(31-575) | BBa_K2201005 | cds with lacUV5 promotor and a strong RBS ( BBa_B0034 ), truncated version lacking the first 30 amino acids |
| pSB1C3-PlacUV5-pelB-SP-PtNTT2 | BBa_K2201006 | cds with lacUV5 promotor and a strong RBS ( BBa_B0034 ), native signal peptide replaced with the pelB signal peptide |
| pSB1C3-PlacUV5-TAT-SP-PtNTT2 | BBa_K2201007 | cds with lacUV5 promotor and a strong RBS ( BBa_B0034 ), native signal peptide replaced with a TAT signal peptide |
| pSB1C3-PlacUV5-PtNTT2-GFP | BBa_K2201002 | Fusion protein of BBa_ K2201000 with GFP ( BBa_E0040 ), cMyc epitope tag as linker ( BBa_K2201181 ) |
| pSB1C3-PlacUV5-PtNTT2(66-575)-GFP | BBa_K2201003 | Fusion protein of BBa_ K2201001 with GFP ( BBa_E0040 ), cMyc epitope tag as linker ( BBa_K2201181 ) |
| pSB1C3-PlacUV5-PtNTT2(31-575)-GFP | BBa_K2201011 | Fusion protein of BBa_K2201005 with GFP ( BBa_E0040 ), cMyc epitope tag as linker ( BBa_K2201181 ) |
| pSB1C3-PlacUV5-pelB-SP-PtNTT2-GFP | BBa_K2201012 | Fusion protein of BBa_K2201006 with GFP ( BBa_E0040 ), cMyc epitope tag as linker ( BBa_K2201181 ) |
| pSB1C3-PlacUV5-TAT-SP-PtNTT2-GFP | BBa_K2201013 | Fusion protein of BBa_K2201007 with GFP ( BBa_E0040 ), cMyc epitope tag as linker ( BBa_K2201181 ) |
Cultivations
Shake Flask Cultivation
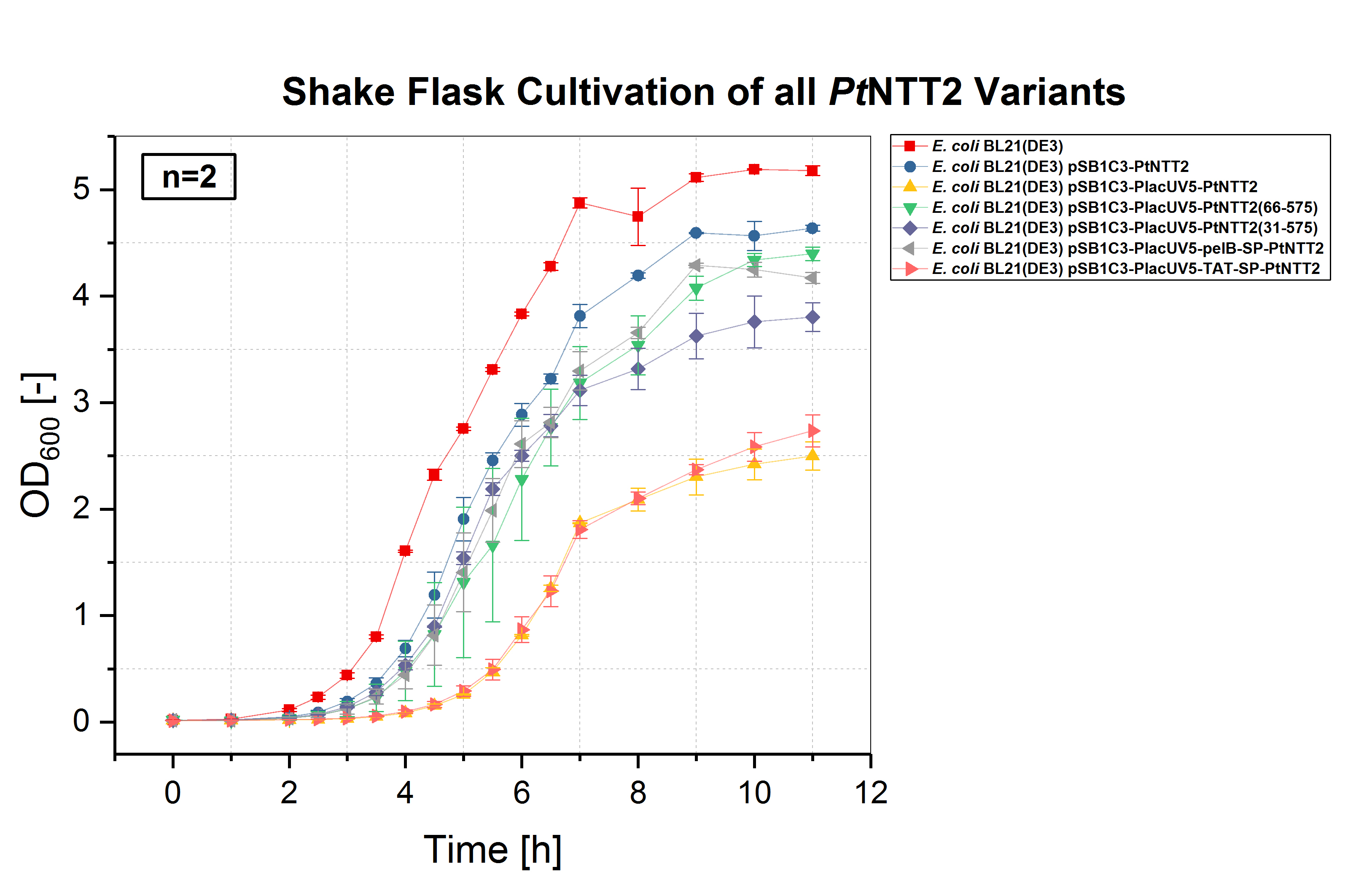
E. coli BL21(DE3) and E. coli BL21(DE3) pSB1C3-PtNTT2, not expressing PtNTT2, were used as negative controls. Two biological replicates of each strain were cultivated and three technical replicates taken for each measurement. A clear difference in the growth rates can be observed, with E. coli BL21(DE3) pSB1C3-PlacUV5-PtNTT2 and E. coli BL21(DE3) pSB1C3-PlacUV5-TAT-SP-PtNTT2 showing the weakest growth. Both strains also show the longest lag phase, which is nearly twice as long as the lag phase of E. coli BL21(DE3). E. coli BL21(DE3) pSB1C3-PlacUV5-PtNTT2(66-575) and E. coli BL21(DE3) pSB1C3-PlacUV5-pelB-SP-PtNTT2 show the best growth of all PtNTT2 variants, reaching the highest OD600.
Given that Zhang et al., 2017 reported some kind of toxicity resulting from the N-terminal sequence of PtNTT2, we first investigated if we could also observe the same toxicity associated with the N-terminal sequence. We also tested whether our own versions of the transporter result in better growth and reduced toxicity compared to the native transporter. Therefore, after cloning the plasmids in E. coli DH5α and verifying the correct assembly via sequencing, all plasmids were transformed into E. coli BL21(DE3). The presence of the correct plasmids was again verified by colony PCR. The first cultivations were carried out in shake flasks in LB media. A total cultivation volume of 50 mL was used. The cultures were incubated at 37 °C and 180 rpm. All cultures were inoculated with an OD600 of 0.01. OD600 was measured every hour during lag and stationary phase and every 30 minutes during the exponential phase. The optical density was measured using an Eppendorf Photometer and standard cuvettes. E. coli BL21(DE3) without a plasmid and E. coli BL21(DE3) harboring pSB1C3-PtNTT2 were used as negative controls. Two biological replicates of each strain were tested and three technical replicates were measured for each timepoint. E. coli BL21(DE3) without any plasmid shows the best growth, with the highest specific growth rate and the highest final OD600 of 5.178 ± 0.046. The second negative control, E. coli BL21(DE3) harboring pSB1C3-PtNTT2 (BBa_K2201004) reached the second highest OD600 with 4.638 ± 0.029. Of the functional PtNTT2 variants, strains harboring pSB1C3-PlacUV5-PtNTT2(66-575) (BBa_K2201001) and pSB1C3-PlacUV5-pelB-SP-PtNTT2 (BBa_K2201006) reached the highest ODs with 4.397 ± 0.062 and 4.171 ± 0.051, respectively. E. coli BL21(DE3) pSB1C3-PlacUV5-PtNTT2(31-575) (BBa_K2201005) showed similar growth to the two previous strains during the lag phase and early exponential phase, but reaching a lower OD600 of 3.802 ± 0.135. E. coli BL21(DE3) pSB1C3-PlacUV5-PtNTT2 (BBa_K2201000) and E. coli BL21(DE3) pSB1C3-PlacUV5-TAT-SP-PtNTT2 (BBa_K2201007) grew significantly weaker compared to all previous strains, with a lag phase nearly four hours long and final ODs of 2.499 ± 0.134 and 2.735 ± 0.150, respectively. The difference in growth between the two negative controls can be explained by the metabolic burden caused by plasmid replication and expression of the chloramphenicol resistance. Therefore, E. coli BL21(DE3) pSB1C3-PtNTT2 is the more accurate control, since all samples contain the same plasmid backbone and were also grown in LB media supplemented with chloramphenicol. The difference in growth between strains expressing the native, full length transporter and the truncated version PtNTT2(66-575), observed by Zhang et al. 2017, could also be shown. This negative effect might be associated with the native signal peptide of PtNTT2, which E. coli might not be able to process correctly. Another explanation for the weak growth could be that the native transporter variant has a higher activity compared to the other variants. If the activity is too high, this might lead to a toxic effect and to the observed weak growth.