Difference between revisions of "Part:BBa K2323002"
| Line 33: | Line 33: | ||
After PCR we ligated the plasmid using the T4 ligase. This sample was then transformed in <i>E. coli</i> DH5&alpha for plasmid storage and <i>E. coli</i> BL21star for protein expression. We expressed the TEV protease in 2xYT medium and purified it via affinity and size exclusion chromatography. | After PCR we ligated the plasmid using the T4 ligase. This sample was then transformed in <i>E. coli</i> DH5&alpha for plasmid storage and <i>E. coli</i> BL21star for protein expression. We expressed the TEV protease in 2xYT medium and purified it via affinity and size exclusion chromatography. | ||
| − | [[Image: | + | |
| + | |||
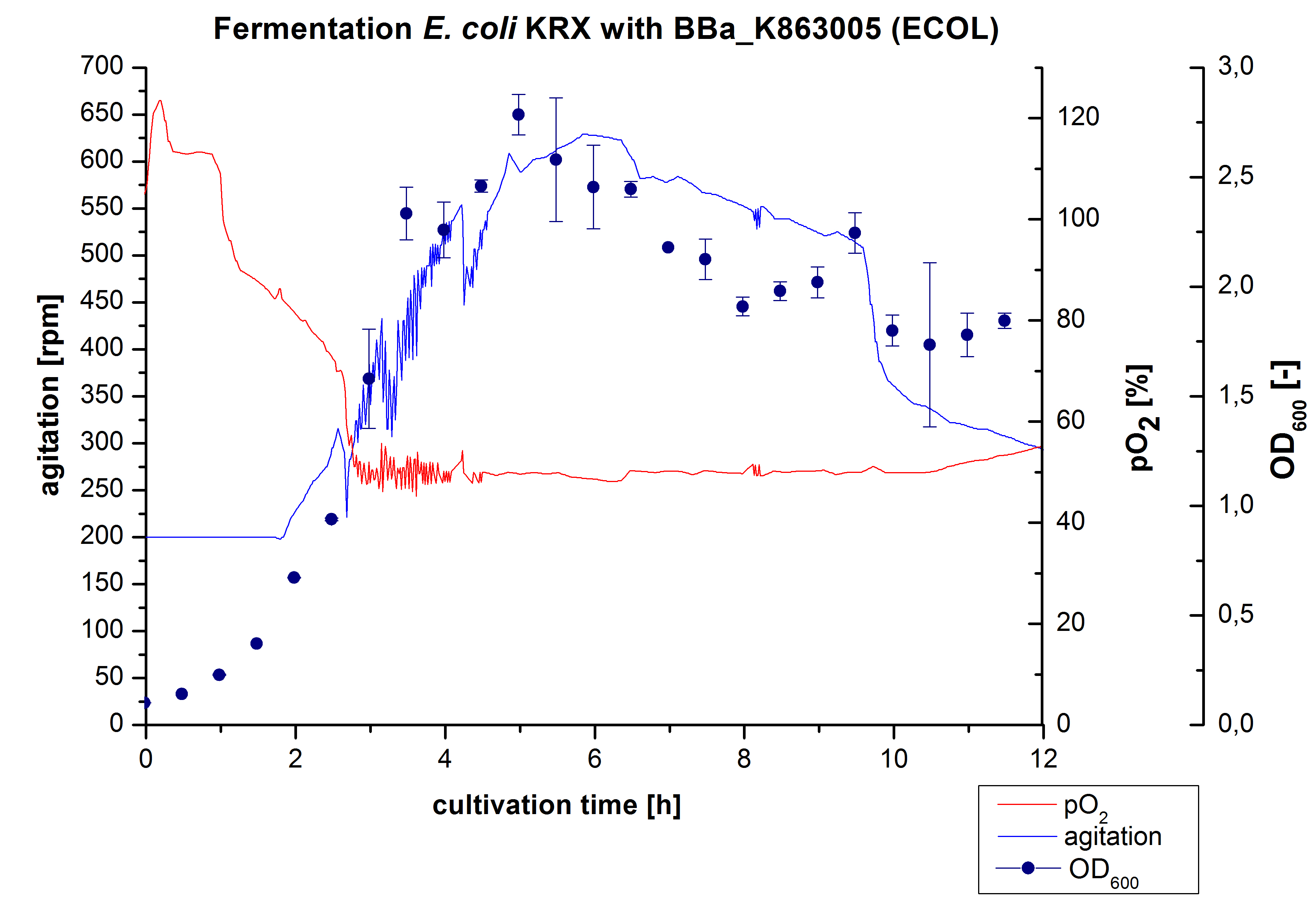
| + | [[Image:Bielefeld2012_ECOL3LFermentation.jpg|450px|thumb|left|'''Figure 1:''' Fermentation of ''E. coli'' KRXwith <partinfo>BBa_K863005</partinfo> (ECOL) in an Infors Labfors Bioreactor, scale: 3 L, [http://2012.igem.org/Team:Bielefeld-Germany/Protocols/Materials#Autoinduction_medium autoinduction medium] + 60 µg/mL chloramphenicol, 37 °C, pH 7, agitation on cascade to hold pO<sub>2</sub> at 50 %, OD<sub>600</sub> measured every 30 minutes.]] | ||
| + | |||
| + | |||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| − | + | [[Image:BBa_K2323002_Plasmid_Map.svg| thumb | 200px | Gel of the Overhang PCR]] | |
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
===Functional Parameters=== | ===Functional Parameters=== | ||
<partinfo>BBa_K2323002 parameters</partinfo> | <partinfo>BBa_K2323002 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
Revision as of 15:16, 29 October 2017
TEV protease with N-terminal 6x His-Tag under the control of the pT7 promoter
Introduction
TEV protease is a highly specific cysteine protease from the Tobacco Etch Virus. An improvement over BBa_K1319008, the protease can be expressed in strains with T7-polymerase and then purified with the help of the His-TAg for synthetic in-vitro circuits.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 71
Illegal AgeI site found at 803 - 1000COMPATIBLE WITH RFC[1000]
Usage and Biology
The (+)-strand RNA genomes are often translated by the host to polyprotein precursors, which are then co-translationally cleaved by therefore provided proteases into the mature proteins. One of these proteases was found in the plant pathogenic Tobacco Etch Virus (TEV). For scientists the TEV protease is a molecular tool to cleave of all sorts of protein tags precisely due to its sequence specificity. It recognizes the amino acid sequence Glu-Asn-Leu-Tyr-Gln-Ser and cleaves then between glutamic acid and serine. In our project, the TEV protease is a main component in the Intein-Extein readout, but also was used in the purification procedure of our Cas13a proteins [http://www.jbc.org/content/277/52/50564.long].
For scientists the TEV protease is a molecular tool to cleave of all sorts of protein tags precisely due to its sequence specificity. It recognizes the amino acid sequence Glu-Asn-Leu-Tyr-Gln-Ser and cleaves then between glutamic acid and serine. In our project, the TEV protease is a main component in the Intein-Extein readout, but also was used in the purification procedure of our Cas13a proteins. We improved the Biobrick BBa_K1319008 by adding a 6x His-tag, which made it possible to purify this protease.
Cloning, expression and Purification
The His-tag was added to pSB1C3-BBa-K1319008 by PCR with overhang primers p-TEV-His-fwd and p-TEV-His-rev.
| Name | 5'-3' primers sequences |
|---|---|
| p-TEV-His-fwd | catcatcaccatcaccacgccggcggcgaaagc |
| p-TEV-His-rev | catctagtatttctcctctttctctagtatctccc |
After PCR we ligated the plasmid using the T4 ligase. This sample was then transformed in E. coli DH5&alpha for plasmid storage and E. coli BL21star for protein expression. We expressed the TEV protease in 2xYT medium and purified it via affinity and size exclusion chromatography.