Difference between revisions of "Part:BBa K2350004"
(→Usage and Biology) |
|||
| Line 3: | Line 3: | ||
== Usage and Biology == | == Usage and Biology == | ||
| − | NrtA is a high-affinity periplasmic solute-binding lipoprotein .It is composed of two domains. Both domains consist of a five-stranded mixed | + | NrtA is a high-affinity periplasmic solute-binding lipoprotein .It is composed of two domains. Both domains consist of a five-stranded mixed sheet surrounded by helices. Nitrate occupies the cleft between the two domains in catching phase. Like most bacterial periplasmic-binding proteins, NrtA is |
tethered to the membrane surface by a lipidated cysteine and a very long flexible linker rich in glycine and serine. Therefore NrtA is akin to a ‘‘balloon on a string’’with its solute-binding domain capturing nitrate/nitrite in the periplasm and transporting it to the NrtB transmembrane permease. | tethered to the membrane surface by a lipidated cysteine and a very long flexible linker rich in glycine and serine. Therefore NrtA is akin to a ‘‘balloon on a string’’with its solute-binding domain capturing nitrate/nitrite in the periplasm and transporting it to the NrtB transmembrane permease. | ||
The order of the strands within domain from some researchers’ point of view suggest that NrtA belongsto class II of the periplasmic-binding protein (PBP) superfamily (7) which also includes the oxyanion-binding proteins. | The order of the strands within domain from some researchers’ point of view suggest that NrtA belongsto class II of the periplasmic-binding protein (PBP) superfamily (7) which also includes the oxyanion-binding proteins. | ||
| Line 12: | Line 12: | ||
same affinity. | same affinity. | ||
We arrange the coding region with a moderate constitutive promoter, and then nrtA will be all over the modified microbe’s cell membrane. | We arrange the coding region with a moderate constitutive promoter, and then nrtA will be all over the modified microbe’s cell membrane. | ||
| + | |||
== Analysis == | == Analysis == | ||
Revision as of 14:06, 27 October 2017
NrtA (a part of nitrate channel protein from Synechocystis sp. PCC 6803)
This part produces a part of nitrate channel protein from Synechocystis sp. PCC 6803 which plays the role of catching nitrate ion from the environment.
Usage and Biology
NrtA is a high-affinity periplasmic solute-binding lipoprotein .It is composed of two domains. Both domains consist of a five-stranded mixed sheet surrounded by helices. Nitrate occupies the cleft between the two domains in catching phase. Like most bacterial periplasmic-binding proteins, NrtA is tethered to the membrane surface by a lipidated cysteine and a very long flexible linker rich in glycine and serine. Therefore NrtA is akin to a ‘‘balloon on a string’’with its solute-binding domain capturing nitrate/nitrite in the periplasm and transporting it to the NrtB transmembrane permease. The order of the strands within domain from some researchers’ point of view suggest that NrtA belongsto class II of the periplasmic-binding protein (PBP) superfamily (7) which also includes the oxyanion-binding proteins. These structures consist of two globular domains of mixed sheet flanked by helices, with the solute-binding cleft located between the two domains. However, NrtA has an extra 100aa extending from the C terminus. These residues comprise several -helices and a two-stranded antiparallel -sheet that cradle the back of the C-clamp. This architectural feature provides structural support for the nitrate-binding pocket or plays a role in the conformational changes associated with solute transport. The resonance state of the nitrate anion in NrtA binding pocket evenly distributes the negative charge among the three oxygen atoms. However, the environment surrounding the nitrate oxygens in the NrtA binding pocket is clearly asymmetric. It seems likely that the O1 and O3 interactions will remain the same but that the O2 atom and its interactions with the protein will be absent in the case of nitrite. O2 in the bound nitrate molecule has relatively few interactions and, hence, maybe the reason why NrtA binds nitrate and nitrite with approximately the same affinity. We arrange the coding region with a moderate constitutive promoter, and then nrtA will be all over the modified microbe’s cell membrane.
Analysis
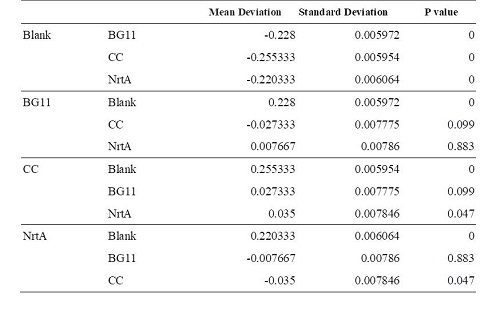
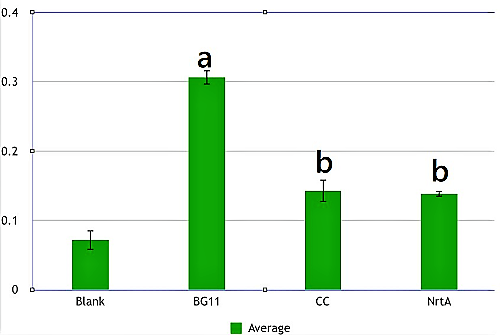
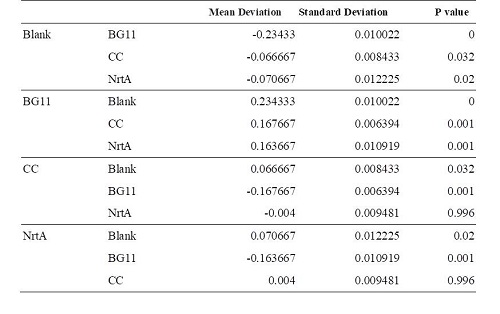
BG11, competent cell, nrtA are not clearly different from each other. However, there is a significant difference between competent cell and nrtA , which represents that nrtA is useful. In addition, after the competent cell is milled , the nitrate , which the ecoli produces on its by itself will be placed inside the medium , which leads to the increasing amount of nitrate.In short, nrtA can combine nitrates as well.
We realize that ecoli, which isn't shattered, has no obvious distinction in the condition of containing/having nrtA and no nrta or not. Yet , both situations will reduce the nitrate.Thus, we may move forward to the goal of transforming the nrta into a secreted protein and releasing it into the medium to absorb the nitrate in the future.
Figure 1. Nitrate absorbance of different cell lysate. Blank:nitrate concentration assay kit assay buffer CC:competent cell BG11:microalgae culture medium buffer NrtA:NrtA producing E.coli.
Table 1. Dunnett 3. Blank:nitrate concentration assay kit assay buffer CC:competent cell BG11:microalgae culture medium buffer NrtA:NrtA producing E.coli.
Figure 2. Nitrate absorbance of cell. Blank:nitrate concentration assay kit assay buffer CC:competent cell BG11:microalgae culture medium buffer NrtA:NrtA producing E.coli.
Table 2. Dunnett 3. Blank:nitrate concentration assay kit assay buffer CC:competent cell BG11:microalgae culture medium buffer NrtA:NrtA producing E.coli
This part produces a part of nitrate channel protein from Synechocystis sp. PCC 6803
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1179
Illegal BamHI site found at 549 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]