Difference between revisions of "Part:BBa K1218011:Experience"
| Line 102: | Line 102: | ||
===BBa_K1218011 could be used to regulate non-coding RNAs=== | ===BBa_K1218011 could be used to regulate non-coding RNAs=== | ||
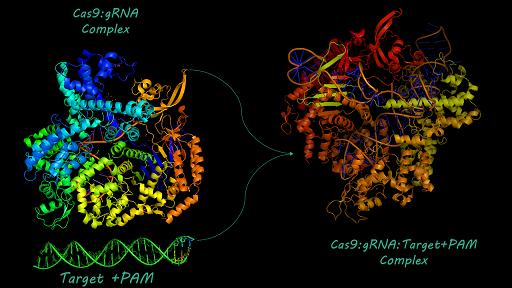
| − | [http://2017.igem.org/Team:AFCM-Egypt# Egypt-AFCM Team] simulated a structural model for [https://parts.igem.org/Part:BBa_K1218011# BBa_K1218011] using SWISS-Model to be ready for protein-nucleic acid docking using HADDOCK algorithm. Docking helped us to have insights about cleavage binding of cas9 to target DNA upstream to PAM site. Docking also revealed mechanisms of interaction between cas9 and gRNA. For more information about [http://2017.igem.org/Team:AFCM-Egypt | + | [http://2017.igem.org/Team:AFCM-Egypt# Egypt-AFCM Team] simulated a structural model for [https://parts.igem.org/Part:BBa_K1218011# BBa_K1218011] using SWISS-Model to be ready for protein-nucleic acid docking using HADDOCK algorithm. Docking helped us to have insights about cleavage binding of cas9 to target DNA upstream to PAM site. Docking also revealed mechanisms of interaction between cas9 and gRNA. For more information about [http://2017.igem.org/Team:AFCM-Egypt# Egypt-AFCM Team] structural Modeling you may visit [http://2017.igem.org/Team:AFCM-Egypt/Model# Egypt-AFCM Modeling] |
Part characterization and usage can be found at this composite part [https://parts.igem.org/wiki/index.php?title=Part:BBa_K2217026# BBa_K2217026_Experience]. | Part characterization and usage can be found at this composite part [https://parts.igem.org/wiki/index.php?title=Part:BBa_K2217026# BBa_K2217026_Experience]. | ||
Revision as of 17:26, 26 October 2017
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Characterization of BBa_K1218011
SKLBC promotes iGEM in Southern China and found two high school software teams SKLBC-China and SKLBC-GDSYZX in 2015. As high school team with the BEST SOFTWARE TOOL prize in iGEM 2014 HS, our team continues to focus on developing software to help iGEMers carry on their experiments and focuses more on software. In order to experience synthetic biology, and further promote the impact of iGEM to our schools and Southern China, this summer, we took the experiment to design a new standard BioBrick Part and characterization of an existing part under the instruction of staff from SKLBC. During the process, we realized that dry-lab and wet-lab could work close together, to bridge the information gap between science and citizens with educational events.
Group: SKLBC-China 2015
Author: SKLBC-China 2015
Group: SKLBC-GDSYZX 2015
Author: SKLBC-GDSYZX 2015
Applications of BBa_K1218011
User Reviews
UNIQdc57d19fbd05ac1a-partinfo-00000000-QINU
|
•••••
CU-Boulder 2014 |
BBa_K1218011 can be targeted to a DNA sequence through the modification of its spacer region.Group: CU-Boulder, 2014 Author: Josephina Hendrix Summary: CU-Boulder demonstrated that BBa_K1218011 can be targeted to a specific DNA sequence through the modification of its spacer region. The original spacer was replaced with one that targeted a neomycin resistance gene. The modified and unmodified plasmids were transformed into cells containing the targeted gene and the decrease in growth with the target sample demonstrates the ability of the spacer to target a specific sequence. Documentation: A 30mer spacer sequence targeting the neomycin phosphotransferase gene was designed and substituted for the original spacer in BBa_K1218011. BW23115 E. coli with the neomycin phosphotransferase inserted into the genome were chemically transformed with the original and modified BBa_K1218011 to compare CRISPR-Cas9 specificity. Transformants were selected for on chloramphenicol. There was a substantial decrease in growth between the non-targeting (1920 colonies) and the targeting sample (8 colonies) that must be accredited to the differences in spacer sequence. As can be seen in Figure 1B), there is growth in the targeting sample. Sequencing showed that all eight colonies had deleted the spacer region and one or both of the adjacent repeats.
Author: Josephina Hendrix Summary: Part BBa_K1218011 was further improved by the addition of the M13 packaging signal (BBa_K1445000) to form the composite part BBa_K1445001. This allows the part to be packaged into M13 phage and delivered to bacteria through phage rather than a transformation method. Documentation:The M13 packaging signal was inserted upstream of the CRISPR-Cas9 part and submitted to the registry as part BBa_K1445001. This new part was packaged into phage and introduced via infection to conjugated neomycin resistant bW23115 E. coli. Sample was plated on chloramphenicol to select for cells infected with the phage delivering the phagemid with the Cas9 part and M13 origin of replication. The growth in figure 2 demonstrates that, with the addition of the M13 origin of replication, BBa_K1218011 can be delivered to cells via recombinant M13 phage.
|
|
•••••
Egypt-AFCM17 |
BBa_K1218011 could be used to regulate non-coding RNAs[http://2017.igem.org/Team:AFCM-Egypt# Egypt-AFCM Team] simulated a structural model for BBa_K1218011 using SWISS-Model to be ready for protein-nucleic acid docking using HADDOCK algorithm. Docking helped us to have insights about cleavage binding of cas9 to target DNA upstream to PAM site. Docking also revealed mechanisms of interaction between cas9 and gRNA. For more information about [http://2017.igem.org/Team:AFCM-Egypt# Egypt-AFCM Team] structural Modeling you may visit [http://2017.igem.org/Team:AFCM-Egypt/Model# Egypt-AFCM Modeling] Part characterization and usage can be found at this composite part BBa_K2217026_Experience. [http://2017.igem.org/Team:AFCM-Egypt# Egypt-AFCM Team] aimed to use cas9 for non-coding RNAs editing represented in regulation of circular RNA based circuit to knock-in hsa_circ_0000064 at BBa_K2217001 into Hepatocellular carcinoma cells. To improve characterization of BBa_K1218011, We designed a CRISPR-based circuit with HDR (homology-directed repair) template for knock-in, to improve characterization CMV enhancer, CMV promoter and T7 promoter at [BBa_K2217000 BBa_K2217006 BBa_K2217007] were ligated to the same circuit with cas9, while HDR was ligated on a separate plasmid to be transfected to HepG2 cells for circuit evaluation. herein, Gel electrophoresis bands were described to document Cas9 characterization, while culture plates were also documented to compare the activity of both non-coding RNA circuit and CRISPR circuit. Information about Our Team results can be found at [http://2017.igem.org/Team:AFCM-Egypt/Results# Egypt-AFCM Team Results]


|
UNIQdc57d19fbd05ac1a-partinfo-00000004-QINU