Difference between revisions of "Part:BBa C0077"
(→Characterisation data of Cin (no LVA tag)) |
(→I. CinR (no LVA) Part by Imperial College London iGEM 2016) |
||
| Line 13: | Line 13: | ||
Group: <b>Imperial College London 2016</b> | Group: <b>Imperial College London 2016</b> | ||
| − | We created a new version of this part without the LVA degradation tag [https://parts.igem.org/Part:BBa_K1893028 (BBa_K1893028)]. | + | We created a new version of this part without the LVA degradation tag [https://parts.igem.org/Part:BBa_K1893028 (BBa_K1893028)]. |
===Characterisation data of Cin (no LVA tag)=== | ===Characterisation data of Cin (no LVA tag)=== | ||
Revision as of 21:00, 28 October 2016
cinR activator from Rhizobium leguminosarum (+LVA)
In complex with O3-C14:1-HSL (made by BBa_C0076), CinR binds to the Cin promoter (BBa_R0077) and activates transcription.
Usage and Biology
In Rhizobium leguminosarum, cinR and cinI are forward-pointing, adjacent, and separate transcriptional elements. There is a short (<200bp) intergenic region between cinR and cinI. The cinR/intergenic/cinI structure is conserved for the cinRI locus in Rhizobium etli, and for the cerRI locus in Rhodobacter sphaeroides. The intergenic region is thought to contain a terminator for cinR, a promoter and RBS for cinI, and a dna-binding domain for cinR and its homologs, though a conserved structure has not yet been identified.
I. CinR (no LVA) Part by Imperial College London iGEM 2016
Group: Imperial College London 2016
We created a new version of this part without the LVA degradation tag (BBa_K1893028).
Characterisation data of Cin (no LVA tag)
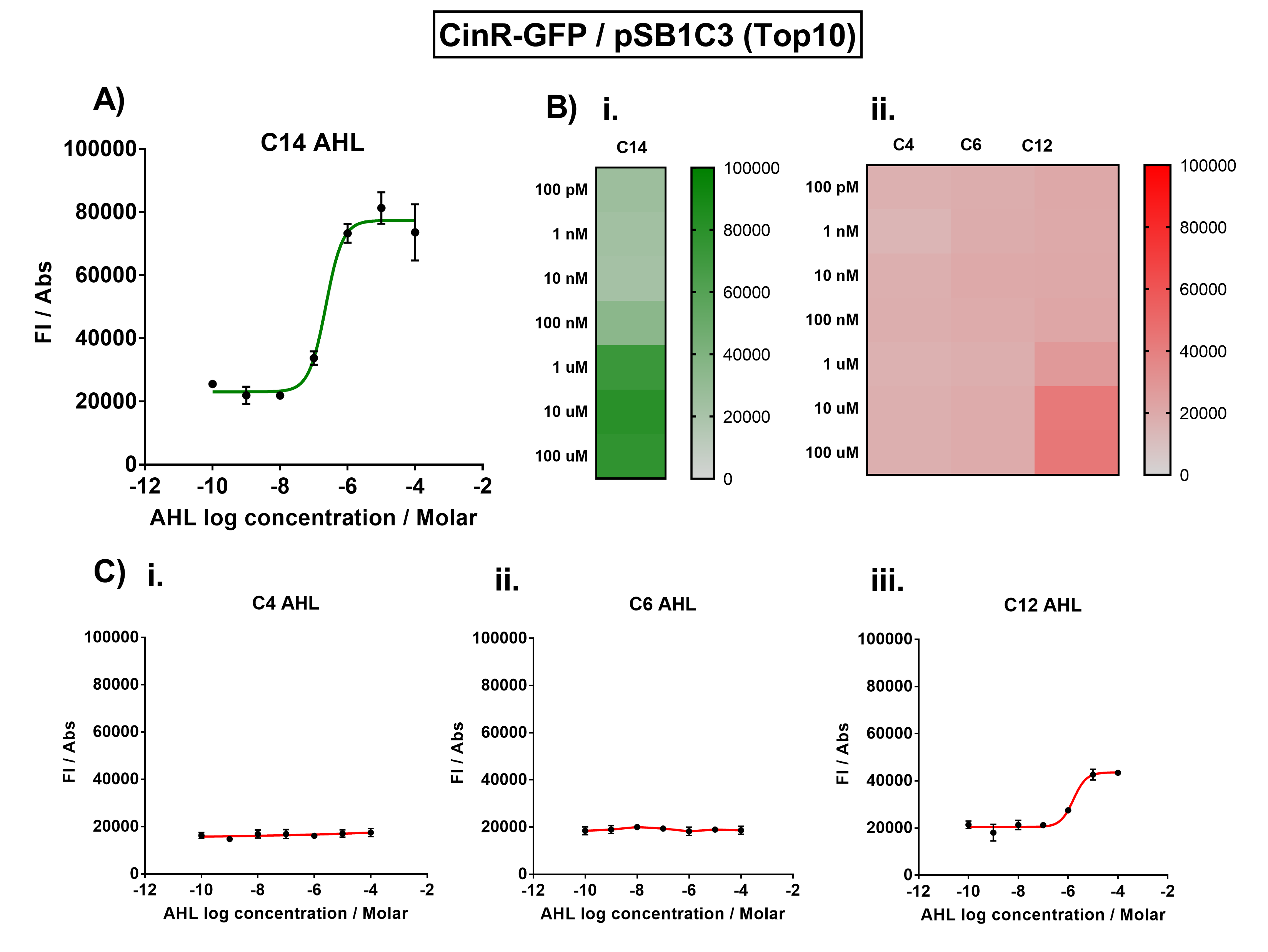
The activation range of CinR by its cognate inducer (3O-C14 AHL) was characterised in TOP10 E.Coli cells. Cells transformed with the Cin response device were cultured to the exponential phase and treated with appropriate concentrations of 3O-C14 AHL in 96-well microplates. These induced cells were grown in the microplates and their fluorescence and absorbance values (OD 600) were monitored over time using a microplate reader. The reported values for the normalised fluorescence represents the values recorded 180 minutes after AHL induction. The normalised fluorescence was calculated by dividing fluorescence values by absorbance values and correcting for LB autofluorescence.
In order to characterise the orthogonality of the Cin system, we measured absorbance and fluorescence of the cells in the plate reader after treating them with varying concentrations of 3 different AHL signals (C4 AHL of the Rhl system, 3O-C6 AHL of the Lux system, and 3O-C12 AHL of the Las system). This allowed us to determine whether these non-cognate AHLs were capable of activating the CinR response protein, and therefore the level of crosstalk between the quorum sensing systems.
Figure 1. Characterisation of the Cin response device (BBa_K1893007). (A) Transfer function curve of normalised fluorescence against cognate inducer C14-AHL concentrations. (B) Heat map of normalised fluorescence of CinR-GFP system over a range of AHL concentrations: (i) Binding of CinR-GFP to its cognate AHL (3O-C14 AHL). (ii) Binding of RhlR-GFP to 3 non-cognate AHLs (C4 AHL, 3O-C6 AHL, 3O-C12 AHL). (C) Transfer function curves of normalised fluorescence against non-cognate inducer AHL (3O-C14 AHL) concentrations to investigate inducer AHL crosstalk: (i) C4-AHL (C4 AHL) of the Rhl system (ii) C6-AHL (3O-C6 AHL) of the Lux system (iii) C12-AHL (3O-C12 AHL) of the Las system. Experiments were performed in E. coli Top10 cell strain cultured at 37°C. Normalised fluorescence was calculated by dividing fluorescent signal by cell density (OD600). Fluorescence measurements were recorded at 180 minutes. Reported values represent the mean normalised fluorescence value from 3 technical repeats and error bars represent standard deviation of these.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 528
- 1000COMPATIBLE WITH RFC[1000]