Difference between revisions of "Part:BBa K2086000"
| Line 17: | Line 17: | ||
===Biology and Our Application=== | ===Biology and Our Application=== | ||
[[Image:T--UNebraska-Lincoln--Biosynthesis.gif|450px|thumb|right|'''Figure 1:''' The serine biosynthesis pathway. Provided by: EcoCyc, a member of the BioCyc database collection http://ecocyc.org/ECOLI/NEW-IMAGE?type=PATHWAY&object=SERSYN-PWY&show-citations=NIL]] | [[Image:T--UNebraska-Lincoln--Biosynthesis.gif|450px|thumb|right|'''Figure 1:''' The serine biosynthesis pathway. Provided by: EcoCyc, a member of the BioCyc database collection http://ecocyc.org/ECOLI/NEW-IMAGE?type=PATHWAY&object=SERSYN-PWY&show-citations=NIL]] | ||
| − | + | The nap operon is a set of genes found in wild type <i>E. coli</i> that codes for the periplasmic nitrate reductases. These proteins comprise one of the pathways native to E. coli that is responsible for reducing nitrate ions to nitrite ions. Although the entire operon contains many genes, this BioBrick contains only the napD, napA, napB, and napC genes. Due to size restrictions in cloning and the fact that the other genes in the operon are not needed to increase the rate of nitrate reduction, we excluded many genes found in the natural operon when creating this BioBrick.</p> | |
| − | <p> | + | <p>The napA and napB genes form the complex that is responsible for the reduction of nitrate to nitrite. The napC protein is a membrane-anchored tetraheme that transfers electrons to the napAB complex, allowing electron flow between membrane and periplasm. The napD protein is responsible for the correct assembly of the napAB enzyme.</p> |
| + | <p>Incorporation of this operon into E. coli should lead to an increase in concentration of the selected nap gene products. This in turn should lead to an increased rate of nitrate reduction. The use of this operon in tandem with other reductases would enable to complete reduction of the nitrate ion to nitrogen gas by E. coli (or another chassis,) a novel functionality for this organism.</p> | ||
| − | + | ===Synthesis of the napDABC operon=== | |
| − | + | <p>Synthesis of the napDABC operon BioBrick was complicated due to the presence of an illegal restriction site as well as the size and difficulty associated with the ligation of non-adjacent regions of the natural operon. The nap operon found in wild type <i>E. coli</i> has the napD and the napA genes adjacent to each other but separated from the napB and napC genes. This means that we had to synthesize the synthetic operon in fragments, rather than a continuous segment. To assemble the synthetic operon, we ordered a fragment of the napDA genes with the BioBrick prefix included and with a silent mutation to eliminate the illegal site. We synthesized the rest of the napA from a wild type strain (MG1655) via PCR using a mutation in the forward primer to eliminate the illegal restriction site. We also used PCR to synthesize the napBC genes from the wild type strain with the only major change being the addition of the BioBrick suffix to the end of the reverse primer. During the process of designing fragments, we ensured that each one had sufficient overlap with those adjacent to it so that they could be easily ligated together. Once all three fragments were successfully obtained, we ligated them using HiFi assembly to create the complete napDABC operon.</p> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | ===Synthesis of | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
===SerA Supplementation=== | ===SerA Supplementation=== | ||
Revision as of 20:47, 19 October 2016
napDABC (Periplasmic Nitrate Reductase)
This contains the NapDABC genes which are believed to be one of the set of genes that are responsible for the reduction of nitrate to nitrite in E. coli.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1991
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 479
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 324
Illegal SapI.rc site found at 238
Illegal SapI.rc site found at 1734
Biology and Our Application
The nap operon is a set of genes found in wild type E. coli that codes for the periplasmic nitrate reductases. These proteins comprise one of the pathways native to E. coli that is responsible for reducing nitrate ions to nitrite ions. Although the entire operon contains many genes, this BioBrick contains only the napD, napA, napB, and napC genes. Due to size restrictions in cloning and the fact that the other genes in the operon are not needed to increase the rate of nitrate reduction, we excluded many genes found in the natural operon when creating this BioBrick.</p>
The napA and napB genes form the complex that is responsible for the reduction of nitrate to nitrite. The napC protein is a membrane-anchored tetraheme that transfers electrons to the napAB complex, allowing electron flow between membrane and periplasm. The napD protein is responsible for the correct assembly of the napAB enzyme.
Incorporation of this operon into E. coli should lead to an increase in concentration of the selected nap gene products. This in turn should lead to an increased rate of nitrate reduction. The use of this operon in tandem with other reductases would enable to complete reduction of the nitrate ion to nitrogen gas by E. coli (or another chassis,) a novel functionality for this organism.
Synthesis of the napDABC operon
Synthesis of the napDABC operon BioBrick was complicated due to the presence of an illegal restriction site as well as the size and difficulty associated with the ligation of non-adjacent regions of the natural operon. The nap operon found in wild type E. coli has the napD and the napA genes adjacent to each other but separated from the napB and napC genes. This means that we had to synthesize the synthetic operon in fragments, rather than a continuous segment. To assemble the synthetic operon, we ordered a fragment of the napDA genes with the BioBrick prefix included and with a silent mutation to eliminate the illegal site. We synthesized the rest of the napA from a wild type strain (MG1655) via PCR using a mutation in the forward primer to eliminate the illegal restriction site. We also used PCR to synthesize the napBC genes from the wild type strain with the only major change being the addition of the BioBrick suffix to the end of the reverse primer. During the process of designing fragments, we ensured that each one had sufficient overlap with those adjacent to it so that they could be easily ligated together. Once all three fragments were successfully obtained, we ligated them using HiFi assembly to create the complete napDABC operon.
SerA Supplementation

Before interpreting the results from our composite part including SerA, it's important to first consider whether our selected auxotroph (JW2880) was able to be rescued with a supplemental SerA gene. By complementing our auxotroph with a plasmid containing SerA and its native promoter, the growth curve in Figure 2 was obtained. The ability of our isolated SerA gene to rescue our auxotroph was confirmed by testing our composite BioBrick kill switch (BBa_K2086002), which expresses the SerA gene when nitrate concentrations reach a certain threshold. The qualitative results from one of these experiments are pictured in Figure 3.
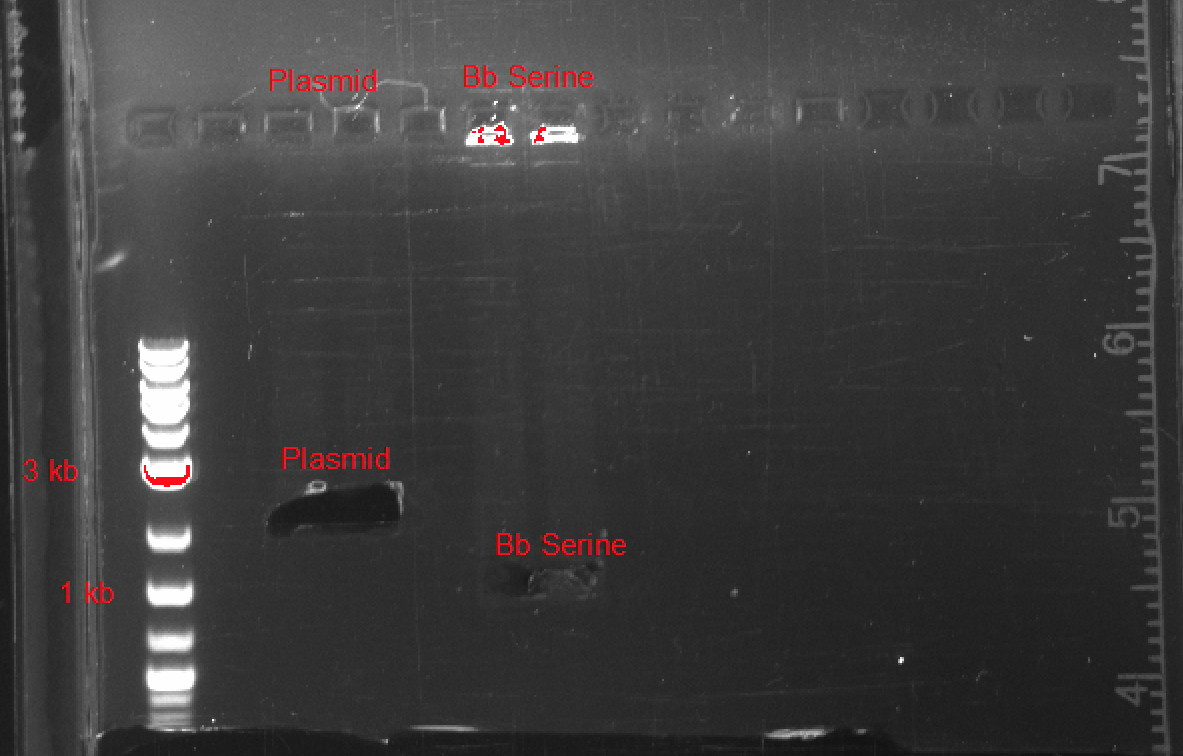
Plasmid Synthesis
After testing, we were able to synthesize a basic SerA ORF plasmid, biobrick compatible with a wide variety of plasmids, promoters, and ribosome binding sites. In Figure 4, you can see the results of our gel electrophoresis, used to ligate the two components of our plasmid:
- pSB1C3 : 2070 bp
- SerA : 1233 bp