Difference between revisions of "Part:BBa K1998004"
SWinchester (Talk | contribs) |
SWinchester (Talk | contribs) |
||
| Line 21: | Line 21: | ||
===Biology & Literature=== | ===Biology & Literature=== | ||
| − | This | + | |
| − | + | PsbM encodes a highly hydrophobic protein, psbM, with a single, membrane spanning α-helix located at the monomer-monomer interface of PSII (Ferreira, Iverson, Maghlaoui, Barber, & Iwata, 2004). This proteins acts to stabilise the dimerization of PSII. While not a necessary factor for PSII biosynthesis, its absence weakens the dimer interconnection of the core complex, and may impair PSII repair (Umate et al., 2007). Deletion studies of this gene have shown a 28% decrease in successfully assembled PSII centres (Bentley, Luo, Dilbeck, Burnap, & Eaton-Rye, 2008). | |
| − | + | ||
| + | PsbZ, also referred to as ycf9, is a highly conserved gene amongst photosynthetic species, encoding a two transmembrane helix protein, psbZ (Ferreira et al., 2004). PsbZ is found at the interface between PSII and light harvesting complex II (LHCII). (Minagawa & Takahashi, 2004). Deletion studies of psbZ have resulted in decreased stability of the PSII-LHCII supercomplex, suggesting this protein is involved in anchoring the two complexes (Swiatek et al., 2001). | ||
| + | |||
| + | PsbH encodes a low molecular weight PSII subunit, psbH, containing multiple phosphorylation sites (Vener, Harms, Sussman, & Vierstra, 2001). Deletion studies of this gene across multiple species have shown different effects. In Synechocystis sp., a slower growth rate, higher light sensitivity, and impaired electron transport from QA to QB has been observed (Mayes et al., 1993). In addition, the deletion of psbH has been observed to both destabilize the PSII complex, and impair the binding of bicarbonate to the complex (Komenda, Lupínková, & Kopecký, 2002), and in Chlamydomonas reinhardtii, eliminate the formation of the PSII complex, revealing psbH as a vital gene for the synthesis of Photosystem II (Summer, Schmid, Bruns, & Schmidt, 1997). | ||
| + | |||
===Protein information=== | ===Protein information=== | ||
| − | + | psbM | |
| − | + | <br> | |
| + | mass: 3.76kDa | ||
| + | <br> | ||
| + | sequence: MEVNIYGLTATALFIIIPTSFLLILYVKTASTQD | ||
| + | <br><br> | ||
| − | + | psbZ | |
| − | + | ||
| − | + | ||
| − | + | ||
<br> | <br> | ||
| + | mass: 4.56kDa | ||
| + | <br> | ||
| + | sequence: MVGVPVVFATPNGWTDNKGAVFSGLSLWLLLVFVVGILNSFVV | ||
| + | <br><br> | ||
| − | + | psbH | |
| − | + | ||
<br> | <br> | ||
| − | + | mass: 6.02kDa | |
<br> | <br> | ||
| − | + | sequence: MSEAGKVLPGWGTTVLMAVFILLFAAFLLIILEIYNSSLILDDVSMSWETLAKVS | |
| + | <br><br> | ||
| + | |||
| + | psbW | ||
<br> | <br> | ||
| + | mass: 9.2kDa | ||
| + | <br> | ||
| + | sequence: MATTVRSEVAKKVAMLSTLPATLAAHPAFALVDERMNGDGTGRPFGVNDPVLGWVLLGVFGTMWAIWFIGQKDLGDFEDADDGLKL | ||
| + | <br><br> | ||
| + | |||
| + | psbK | ||
| + | <br> | ||
| + | mass: 5.0kDa | ||
| + | <br> | ||
| + | sequence: MTTLALVLAKLPEAYAPFAPIVDVLPVIPVFFILLAFVWQAAVSFR | ||
| + | <br><br> | ||
| + | |||
| + | ===References=== | ||
| + | [1] | ||
| + | |||
| + | [2] | ||
| + | |||
| + | [3] | ||
| + | |||
| + | [4] | ||
| + | |||
| + | [5] | ||
Revision as of 04:20, 18 October 2016
psbMZHWK
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 570
- 1000COMPATIBLE WITH RFC[1000]
Overview
This part is composed of the psbM, psbZ, psbH, psbW and psbK genes. The psbM protein subunit is positioned at the monomer-monomer interface. The psbZ protein controls the interaction of Photosystem II cores with the light-harvesting antenna. The psbH protein is required for stability and assembly of the photosystem II complex. The psbW protein stabilizes dimeric photosytem II. The psbK protein is also required for stability and assembly of Photosystem II.
These parts make up one of the operons in our PSII pathway.
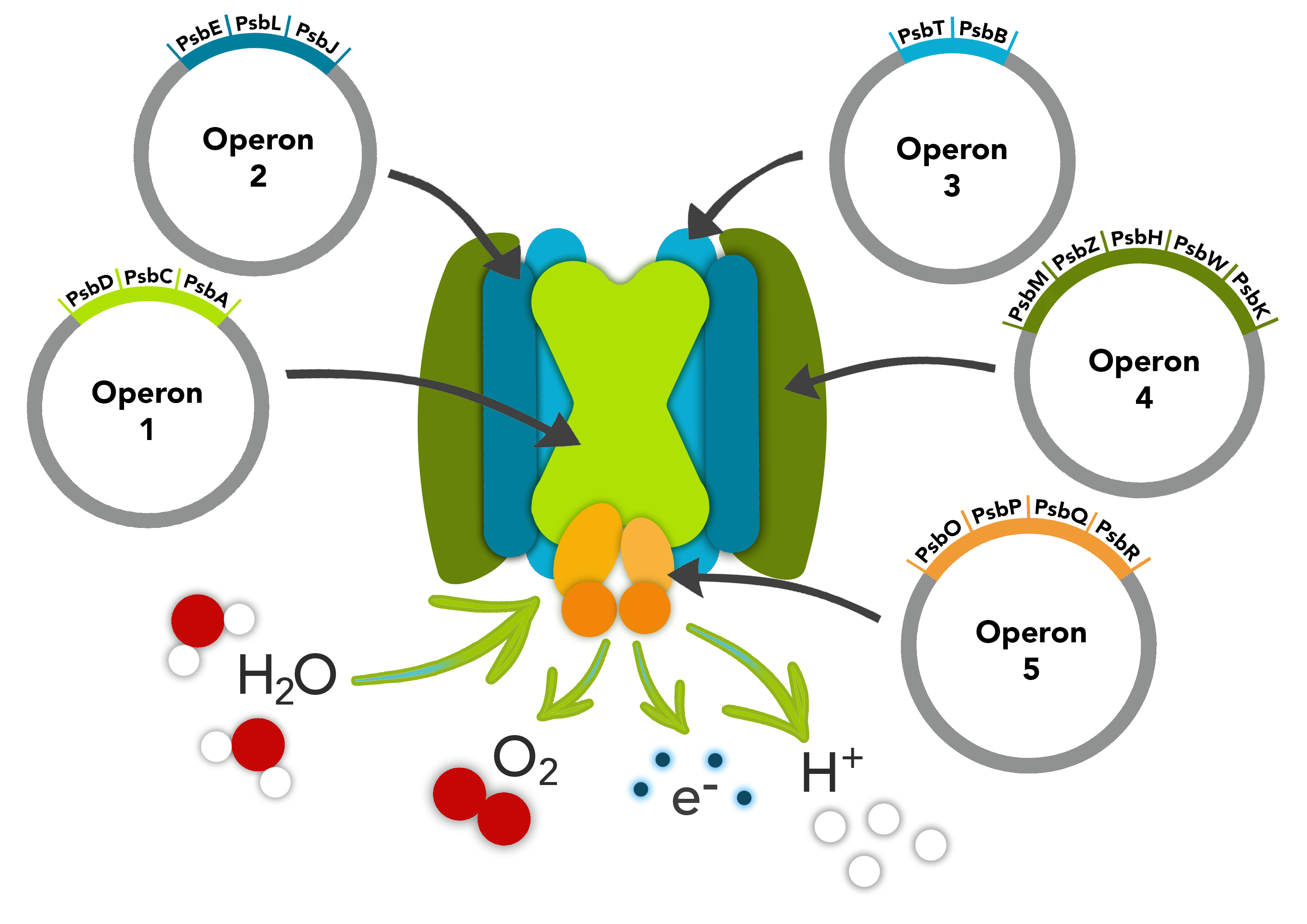
Biology & Literature
PsbM encodes a highly hydrophobic protein, psbM, with a single, membrane spanning α-helix located at the monomer-monomer interface of PSII (Ferreira, Iverson, Maghlaoui, Barber, & Iwata, 2004). This proteins acts to stabilise the dimerization of PSII. While not a necessary factor for PSII biosynthesis, its absence weakens the dimer interconnection of the core complex, and may impair PSII repair (Umate et al., 2007). Deletion studies of this gene have shown a 28% decrease in successfully assembled PSII centres (Bentley, Luo, Dilbeck, Burnap, & Eaton-Rye, 2008).
PsbZ, also referred to as ycf9, is a highly conserved gene amongst photosynthetic species, encoding a two transmembrane helix protein, psbZ (Ferreira et al., 2004). PsbZ is found at the interface between PSII and light harvesting complex II (LHCII). (Minagawa & Takahashi, 2004). Deletion studies of psbZ have resulted in decreased stability of the PSII-LHCII supercomplex, suggesting this protein is involved in anchoring the two complexes (Swiatek et al., 2001).
PsbH encodes a low molecular weight PSII subunit, psbH, containing multiple phosphorylation sites (Vener, Harms, Sussman, & Vierstra, 2001). Deletion studies of this gene across multiple species have shown different effects. In Synechocystis sp., a slower growth rate, higher light sensitivity, and impaired electron transport from QA to QB has been observed (Mayes et al., 1993). In addition, the deletion of psbH has been observed to both destabilize the PSII complex, and impair the binding of bicarbonate to the complex (Komenda, Lupínková, & Kopecký, 2002), and in Chlamydomonas reinhardtii, eliminate the formation of the PSII complex, revealing psbH as a vital gene for the synthesis of Photosystem II (Summer, Schmid, Bruns, & Schmidt, 1997).
Protein information
psbM
mass: 3.76kDa
sequence: MEVNIYGLTATALFIIIPTSFLLILYVKTASTQD
psbZ
mass: 4.56kDa
sequence: MVGVPVVFATPNGWTDNKGAVFSGLSLWLLLVFVVGILNSFVV
psbH
mass: 6.02kDa
sequence: MSEAGKVLPGWGTTVLMAVFILLFAAFLLIILEIYNSSLILDDVSMSWETLAKVS
psbW
mass: 9.2kDa
sequence: MATTVRSEVAKKVAMLSTLPATLAAHPAFALVDERMNGDGTGRPFGVNDPVLGWVLLGVFGTMWAIWFIGQKDLGDFEDADDGLKL
psbK
mass: 5.0kDa
sequence: MTTLALVLAKLPEAYAPFAPIVDVLPVIPVFFILLAFVWQAAVSFR
References
[1]
[2]
[3]
[4]
[5]
