Difference between revisions of "Part:BBa K1919300"
| Line 20: | Line 20: | ||
<!-- --> | <!-- --> | ||
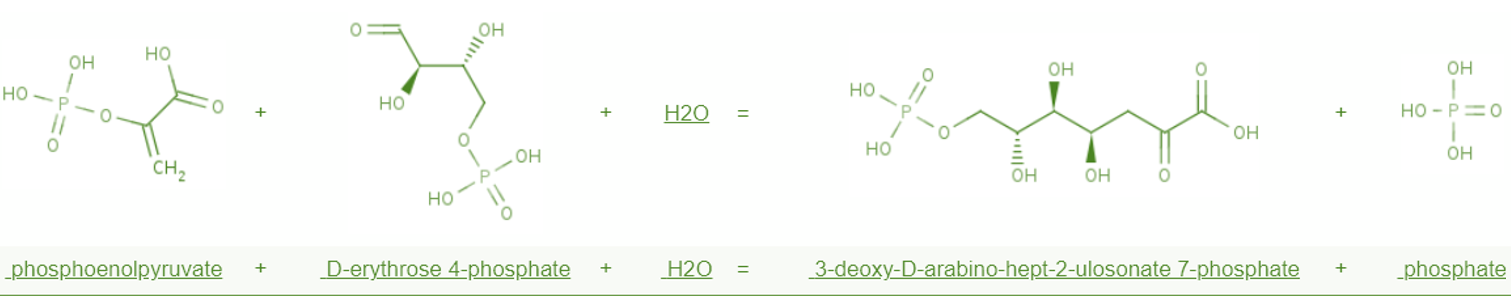
==Quantitative PCR exams expression levels of liv operon== | ==Quantitative PCR exams expression levels of liv operon== | ||
| − | Liv operon (leucine isoleucine valine) contains six genes. We cloned all those genes into | + | Liv operon (leucine isoleucine valine) contains six genes. We cloned all those genes into overexpress vectors in which livJKHMGF are under control of promoter J23100. All those devices have been transferred into E.coli and RNA expression levels are scrutinized via qPCR. In the trial, we regarded two genes, rrsA and dnaA, as reference. The rrsA gene transcripts 16s RNA, while dnaA encodes a component of DNA polymerase. The expression level of rrsA is very high, whereas the dnaA expression level is quite low. Generally speaking, the expression level of gene we need to study is always higher than dnaA and lower than rrsA. The two reference genes expression level are relatively stable. |
[[File:Figure1.png|450px|thumb|left|'''Figure 1:''' the physical position of six genes on genome of E.coli K12.]] | [[File:Figure1.png|450px|thumb|left|'''Figure 1:''' the physical position of six genes on genome of E.coli K12.]] | ||
| − | This figure shows all six genes’ physical position in liv operon. Those six genes participate directly in branched chain amino acid transport. Two periplasmic amino acid binding protein are encoded by livJ and livK. These two proteins confer specificity on LIV-I and LS transport system. The livJ gene product binds leucine, isoleucine and valine, whereas the livK gene product is specific to leucine. These two | + | This figure shows all six genes’ physical position in liv operon. Those six genes participate directly in branched chain amino acid transport. Two periplasmic amino acid binding protein are encoded by livJ and livK. These two proteins confer specificity on LIV-I and LS transport system. The livJ gene product binds leucine, isoleucine and valine, whereas the livK gene product is specific to leucine. These two systems share a set of membrane components, products of the livHMGF genes. LivH and livM are estimated two transmembrane proteins locateing in innermenbrane of E.coli. An analysis of the livH and livM DNA sequence suggests that they encode hydrophobic proteins capable of spanning the membrane several times. The livG and livF proteins are less hydrophobic, but are also tightly associated with the membrane. Both livG and livF contain the consensus sequence for adenine nucleotide binding observed in many other transport proteins. |
</p> | </p> | ||
| Line 36: | Line 36: | ||
<p align="justify"> | <p align="justify"> | ||
| − | [[File:Figure 2.png|450px|thumb|left|'''Figure 2:''' we presumed the folds of expression level of rrsA over dnaA are 10^10. Then the six genes expression | + | [[File:Figure 2.png|450px|thumb|left|'''Figure 2:''' we presumed the folds of expression level of rrsA over dnaA are 10^10. Then the six genes expression levels are calculated according to reference gene.]] |
[[File:Figure3.png|450px|thumb|left|'''Figure 3:''' the data from figure 1 are collected to show again except for livJ. Since expression level of livJ is so high that other expression level of genes are difficult to look. ]] | [[File:Figure3.png|450px|thumb|left|'''Figure 3:''' the data from figure 1 are collected to show again except for livJ. Since expression level of livJ is so high that other expression level of genes are difficult to look. ]] | ||
| − | The figure 2 and 3 show different expression level of six genes compared to dnaA after transferring six | + | The figure 2 and 3 show different expression level of six genes compared to dnaA after transferring six overexpress vectors into DH5a and BL21 when presuming the folds of expression level of rrsA over dnaA are 10^10, that is, we normalized the folds of expression levels of rrsA over dnaA in BL21 and DH5a to 10^10. Usually, normalization helps us to compare different levels under the same background. Please note that the number in vertical axis do not reflects reality. |
</p> | </p> | ||
| Line 47: | Line 47: | ||
<br style="clear: both" /> | <br style="clear: both" /> | ||
| − | ===data from reference genes suggested folds of expression | + | ===data from reference genes suggested folds of expression levels of rrsA over dnaA in BL21 and DH5a are distinct=== |
<p align="justify"> | <p align="justify"> | ||
| − | [[File:Figure 4.png|450px|thumb|left|'''Figure 4:''' different expression level folds of rrsA over dnaA | + | [[File:Figure 4.png|450px|thumb|left|'''Figure 4:''' different expression level folds of rrsA over dnaA in DH5a and BL21. T-test P<0.01 ]] |
| − | We found that expression level folds of rrsA over dnaA are very distinct (T-test, P<0.01) | + | We found that expression level folds of rrsA over dnaA in BL21 and DH5a are very distinct (T-test, P<0.01). The expression level of rrsA in BL21 is around 160,000 times higher than that of dnaA, while it is around 100,000 times in DH5a. Therefore, we take the distinct expression folds of rrsA and dnaA into consideration. |
</p> | </p> | ||
| Line 60: | Line 60: | ||
<br style="clear: both" /> | <br style="clear: both" /> | ||
| − | ===data process 2: take discrepancy of folds of expression level of rrsA over dnaA into consideration=== | + | ===data process 2: take discrepancy of folds of expression level of rrsA over dnaA in BL21 and DH5a into consideration=== |
<p align="justify"> | <p align="justify"> | ||
| − | [[File:Figure 5.png|450px|thumb|left|'''Figure 5:''' recalculate expression level after taking distinct expression level folds of rrsA over dnaA into consideration. ]] | + | [[File:Figure 5.png|450px|thumb|left|'''Figure 5:''' recalculate expression level after taking distinct expression level folds of rrsA over dnaA in BL21 and DH5a into consideration. ]] |
[[File:Figure 6.png|450px|thumb|left|'''Figure 6:''' the data from figure 5 is showed again except for livJ since the livJ expression level is so high that overshadows others. In this figure, we can clearly recognize other genes’ expression level. ]] | [[File:Figure 6.png|450px|thumb|left|'''Figure 6:''' the data from figure 5 is showed again except for livJ since the livJ expression level is so high that overshadows others. In this figure, we can clearly recognize other genes’ expression level. ]] | ||
| − | Please note that negative control set are not shown in the figure, since all of expression levels of six genes in wild BL21 or DH5a are below 1 in the figure. Albeit the DH5a owns livJKHMGF in genome, the | + | Please note that negative control set are not shown in the figure, since all of expression levels of six genes in wild BL21 or DH5a are below 1 in the figure. Albeit the DH5a owns livJKHMGF in genome, the express of them are repressed by a regulator, Lrp (leucine-responsive regulator protein). The number in vertical axis indicates how many times the expression level of certain gene is higher than that of dnaA. If we assume that the expression level of danA is 1 in BL21 and DH5a, the expression level of livJ, for example, is 2339 in DH5a and 9304 in BL21. As we can see from figure 4 and figure 5, livJ and livK are the two highest expressed genes, especially for livJ. And there is a tendency that expression level of genes reduces in accord with physical position on genome from left to right. We believe it reflects the function of those genes to some extent, since there need more periplasmic binding protein (livJK) than receptors (livHMGF) on membrane so that uptake of amino acid will be more efficient. |
</p> | </p> | ||
| Line 82: | Line 82: | ||
To exam which overexpression vector can promote leu uptake most drastically, we transferred all six genes into BL21 and DH5a. We designed a novel protocol to exam fluctuation of leu concentration in liquid LB medium. The protocol goes that: | To exam which overexpression vector can promote leu uptake most drastically, we transferred all six genes into BL21 and DH5a. We designed a novel protocol to exam fluctuation of leu concentration in liquid LB medium. The protocol goes that: | ||
====Materials that should be prepared in advance==== | ====Materials that should be prepared in advance==== | ||
| − | * 1 sulfosalicylic acid solution 20 g solid sulfosalicylic acid | + | * 1 sulfosalicylic acid solution 20 g solid sulfosalicylic acid is dissolved in 100ml ddwater |
* 2 ninhydrin solution 1 g ninhydrin is dissolved in 35ml ddwater | * 2 ninhydrin solution 1 g ninhydrin is dissolved in 35ml ddwater | ||
* 3 pH8.03 PBS | * 3 pH8.03 PBS | ||
| Line 95: | Line 95: | ||
* 3 pipette 300 ml LB to other two new 1.5 ml tubes and add 100ul sulfosalicylic acid solution | * 3 pipette 300 ml LB to other two new 1.5 ml tubes and add 100ul sulfosalicylic acid solution | ||
* 4 shaking violently for 1 min | * 4 shaking violently for 1 min | ||
| − | * 5 20000g*10min and then pipette 300ul supernatant to a new 1.5 EP tube | + | * 5 20000g*10min for centrifugation and then pipette 300ul supernatant to a new 1.5 EP tube |
* 6 add 1ml pH8.4 PBS buffer and mix them by reversing the tube several times | * 6 add 1ml pH8.4 PBS buffer and mix them by reversing the tube several times | ||
* 7 all 0.15ml ninhydrin solution and mix them by reversing the tube several times | * 7 all 0.15ml ninhydrin solution and mix them by reversing the tube several times | ||
| Line 109: | Line 109: | ||
[[File:Figure 7.png|450px|thumb|left|'''Figure 7:''' full wavelength scanning of liquid LB implemented with leucine (A) or without leucine (B). We detected that there is a high peak of absorption in 570 nm. ]] | [[File:Figure 7.png|450px|thumb|left|'''Figure 7:''' full wavelength scanning of liquid LB implemented with leucine (A) or without leucine (B). We detected that there is a high peak of absorption in 570 nm. ]] | ||
| − | Before further test, we exam two kinds of liquid LB medium that are with or without additional leucine | + | Before further test, we exam two kinds of liquid LB medium that are with or without additional leucine implementation. The liquid medium was processed follow the protocol above and tested by spectrophotometer for full wavelength scanning from 300nm to 1000nm. Since liquid LB medium teems with amino acid, short peptide and other nutrition, we can see a peak in B (without leucine implementation) at 570nm as well. |
<br style="clear: both" /> | <br style="clear: both" /> | ||
| Line 118: | Line 118: | ||
[[File:Figure 8.png|450px|thumb|left|'''Figure 8:''' liquid mediums shaking with DH5a harboring different overexpress vectors are tested by spectrophotometer at 570nm. ]] | [[File:Figure 8.png|450px|thumb|left|'''Figure 8:''' liquid mediums shaking with DH5a harboring different overexpress vectors are tested by spectrophotometer at 570nm. ]] | ||
| − | The optical density as 570nm of liquid LB implemented with leucine. Nctr is a negative control set that do not contain microbe. 2101 is a negative control as well but inoculates with an E.coli named 2101 that harbors chloromycetin resistance but only expresses a short non-coding RNA. LivJKHMGF indicates medium after shaking overnight with E.coli DH5a contain corresponding | + | The optical density as 570nm of liquid LB implemented with leucine. Nctr is a negative control set that do not contain microbe. 2101 is a negative control as well but inoculates with an E.coli named 2101 that harbors chloromycetin resistance but only expresses a short non-coding RNA. LivJKHMGF in the figure indicates medium after shaking overnight with E.coli DH5a contain corresponding overexpress vectors. The data of BL21 is not shown here since the data of different sets only owns tiny discrepency. |
| + | |||
We try to explain these results, but we are not sure the following explanation reflects reality to what extent. To ascertain this question, there need more sophisticated investigation. | We try to explain these results, but we are not sure the following explanation reflects reality to what extent. To ascertain this question, there need more sophisticated investigation. | ||
| + | |||
As we have mentioned before, liv JKHMGF are not resides on genome of Bl21. In addition, the leucine ABC transporter will not work if there is not other component expressed in cell. Thus, it can be understood that there are tiny discrepency among different sets of experiments in BL21. | As we have mentioned before, liv JKHMGF are not resides on genome of Bl21. In addition, the leucine ABC transporter will not work if there is not other component expressed in cell. Thus, it can be understood that there are tiny discrepency among different sets of experiments in BL21. | ||
| − | For DH5a, Albeit the DH5a owns livJKHMGF in genome, the expressions of them are repressed by a regulator, Lrp (leucine-responsive regulator protein). According to the result of BLAST, Lrp not only resides on genome of DH5a but that of BL21. Lrp is a dual transcriptional regulator for at least 10% of the genes in Escherichia coli. These genes are involved in amino acid biosynthesis and catabolism, nutrient transport, pili synthesis, and other cellular functions. It is believed that Lrp senses the presence of rich nutrition based on the concentration of leucine and positively regulates genes that function during famine and negatively regulates genes that function during a feast. Lrp-leucine DNA bingding transcriptional dual regulator inhibits livJ transcription initiation, whereas L-leucine binds to Lrp transcriptional dual regulator to block inhibition of livKHGMF. | + | |
| − | As shows in figure 8, livJK seem do not increase uptake of leucine compared with 2101, livHM inhibit uptake of leucine compared with 2101 and livGF promote uptake of leucine dramatically, especially for livF. We can brace these six genes into 3 group, livJK, livHM and livGF, according to the result of spectrophotometer. Then we attempt to ascertain the inner relationship that brace these genes into the 3 group. | + | For DH5a, Albeit the DH5a owns livJKHMGF in genome, the expressions of them are repressed by a regulator, Lrp (leucine-responsive regulator protein). According to the result of BLAST, Lrp not only resides on genome of DH5a but that of BL21. Lrp is a dual transcriptional regulator for at least 10% of the genes in Escherichia coli. These genes are involved in amino acid biosynthesis and catabolism, nutrient transport, pili synthesis, and other cellular functions. It is believed that Lrp senses the presence of rich nutrition based on the concentration of leucine and positively regulates genes that function during famine and negatively regulates genes that function during a feast. |
| + | |||
| + | Lrp-leucine DNA bingding transcriptional dual regulator inhibits livJ transcription initiation, whereas L-leucine binds to Lrp transcriptional dual regulator to block inhibition of livKHGMF. | ||
| + | |||
| + | As shows in figure 8, livJK seem do not increase uptake of leucine compared with 2101, livHM inhibit uptake of leucine compared with 2101 and livGF promote uptake of leucine dramatically, especially for livF, compared with 2101. We can brace these six genes into 3 group, livJK, livHM and livGF, according to the result of spectrophotometer. Then we attempt to ascertain the inner relationship that brace these genes into the 3 group. | ||
| Line 130: | Line 136: | ||
| − | === | + | ===effect of leucine in coding region=== |
[[File:Figure 9.png|450px|thumb|left|'''Figure 9:''' number of leucine in these six genes. ]] | [[File:Figure 9.png|450px|thumb|left|'''Figure 9:''' number of leucine in these six genes. ]] | ||
| Line 150: | Line 156: | ||
[[File:Figure 13.jpeg|450px|thumb|left|'''Figure 13:'''hydrophilicity plot of livGF ]] | [[File:Figure 13.jpeg|450px|thumb|left|'''Figure 13:'''hydrophilicity plot of livGF ]] | ||
| − | As figure | + | As figure 11,12,13 shows, livJK owns a hydrophobic N terminal. It is the signal peptide that guides livJ and livK to be transported through inner membrane and to be trapped in periplasmic space. livHM should be transmembrane protein since they have very hydrophobic peptide span. LivGF do not contain obvious hydrophobic peptide but contain Walker A and Walker B motifs involved in the binding and hydrolysis of ATP (according to results of BLAST). Thus we can imagine the structure of leucine ABC transporter which is shown in figure 14. |
<br style="clear: both" /> | <br style="clear: both" /> | ||
| Line 160: | Line 166: | ||
[[File:Figure 14.jpeg|450px|thumb|left|'''Figure 14:'''predicted structure of leucine ABC transporter.]] | [[File:Figure 14.jpeg|450px|thumb|left|'''Figure 14:'''predicted structure of leucine ABC transporter.]] | ||
| − | + | Overexpress livJK in DH5a will not increase uptake of leucine in high leucine concentration medium (around 1ug/ml). Overexpress livMH in DH5a inhibits leucine uptake of leucine in high leucine concentration medium (around 1ug/ml). Overexpress livFG in DH5a promotes leucine uptake of leucine in high leucine concentration medium (around 1ug/ml). It may be explained that livFG works like a pump to hydrolysis ATP and obtain energy to transport leucine from periplasmic space into cytosol. More livFG means more energy is used to pump leucine into cytosol. | |
Overexpress livHM may result in more transmembrane pathway which may be a handicap to retain leucine in cell cytosol without futher energy to determine direction of transportation, that is, there will be some leak of leucine when overexpress livHM. | Overexpress livHM may result in more transmembrane pathway which may be a handicap to retain leucine in cell cytosol without futher energy to determine direction of transportation, that is, there will be some leak of leucine when overexpress livHM. | ||
| − | Overexpress livJK can not increase uptake of leucine may be explained that livJ and livK are saturated for transporter livHMGF, that is, more livJK to pass leucine to transporter livHMGF than the transporter needs. In addition, overexpressed livJK do not have enough time to be transported into periplasmic space and congregated | + | Overexpress livJK can not increase uptake of leucine may be explained that livJ and livK are saturated for transporter livHMGF, that is, more livJK to pass leucine to transporter livHMGF than the transporter needs. In addition, overexpressed livJK do not have enough time to be transported into periplasmic space and congregated together in cell and livJK do not have enough enzyme to cleave N-terminal of livJK, which may be another explanation for performance of overexpressed livJK. The two explanation for livJK are not mutual excluded. |
'''Again,please note that all of those explanation of leucine uptake derived from conjecture. To ascertain the puzzle results, further and sophisticated experiment should be carried out.''' | '''Again,please note that all of those explanation of leucine uptake derived from conjecture. To ascertain the puzzle results, further and sophisticated experiment should be carried out.''' | ||
Revision as of 13:33, 17 October 2016
livJ
LivJ encodes periplasmic binding protein for branched-chain amino acid ABC transporter which is encodes by operon livHMGF. The livJ gene product binds leucine, isoleucine, and valine. Please note that there is a short guide peptide on N terminal of livJ coding region, which is used to guide the precusor to be transported into periplasmic room. This device constructed in our team contains a promoter J23100, a RBS, livJ coding region and a terminator. The device can be transferred into E.coli and overexpress livJ directly. The expression of the livJ RNA in the part has been studied via qPCR.The quantified result suggests that compared with negtive control of E.coli (not cantain the plasmid), livJ has been highly expressed in E.coli BL21 and E.coli DH5a.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 382
Illegal AgeI site found at 203
Illegal AgeI site found at 485 - 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 366
Quantitative PCR exams expression levels of liv operon
Liv operon (leucine isoleucine valine) contains six genes. We cloned all those genes into overexpress vectors in which livJKHMGF are under control of promoter J23100. All those devices have been transferred into E.coli and RNA expression levels are scrutinized via qPCR. In the trial, we regarded two genes, rrsA and dnaA, as reference. The rrsA gene transcripts 16s RNA, while dnaA encodes a component of DNA polymerase. The expression level of rrsA is very high, whereas the dnaA expression level is quite low. Generally speaking, the expression level of gene we need to study is always higher than dnaA and lower than rrsA. The two reference genes expression level are relatively stable.
This figure shows all six genes’ physical position in liv operon. Those six genes participate directly in branched chain amino acid transport. Two periplasmic amino acid binding protein are encoded by livJ and livK. These two proteins confer specificity on LIV-I and LS transport system. The livJ gene product binds leucine, isoleucine and valine, whereas the livK gene product is specific to leucine. These two systems share a set of membrane components, products of the livHMGF genes. LivH and livM are estimated two transmembrane proteins locateing in innermenbrane of E.coli. An analysis of the livH and livM DNA sequence suggests that they encode hydrophobic proteins capable of spanning the membrane several times. The livG and livF proteins are less hydrophobic, but are also tightly associated with the membrane. Both livG and livF contain the consensus sequence for adenine nucleotide binding observed in many other transport proteins.
</p>
data process 1: presume folds of expression level of rrsA over dnaA are same in BL21 and in DH5a
The figure 2 and 3 show different expression level of six genes compared to dnaA after transferring six overexpress vectors into DH5a and BL21 when presuming the folds of expression level of rrsA over dnaA are 10^10, that is, we normalized the folds of expression levels of rrsA over dnaA in BL21 and DH5a to 10^10. Usually, normalization helps us to compare different levels under the same background. Please note that the number in vertical axis do not reflects reality.
data from reference genes suggested folds of expression levels of rrsA over dnaA in BL21 and DH5a are distinct
We found that expression level folds of rrsA over dnaA in BL21 and DH5a are very distinct (T-test, P<0.01). The expression level of rrsA in BL21 is around 160,000 times higher than that of dnaA, while it is around 100,000 times in DH5a. Therefore, we take the distinct expression folds of rrsA and dnaA into consideration.
data process 2: take discrepancy of folds of expression level of rrsA over dnaA in BL21 and DH5a into consideration
Please note that negative control set are not shown in the figure, since all of expression levels of six genes in wild BL21 or DH5a are below 1 in the figure. Albeit the DH5a owns livJKHMGF in genome, the express of them are repressed by a regulator, Lrp (leucine-responsive regulator protein). The number in vertical axis indicates how many times the expression level of certain gene is higher than that of dnaA. If we assume that the expression level of danA is 1 in BL21 and DH5a, the expression level of livJ, for example, is 2339 in DH5a and 9304 in BL21. As we can see from figure 4 and figure 5, livJ and livK are the two highest expressed genes, especially for livJ. And there is a tendency that expression level of genes reduces in accord with physical position on genome from left to right. We believe it reflects the function of those genes to some extent, since there need more periplasmic binding protein (livJK) than receptors (livHMGF) on membrane so that uptake of amino acid will be more efficient.
Leu absorption trial
novel protocol for leucine concentration examination
To exam which overexpression vector can promote leu uptake most drastically, we transferred all six genes into BL21 and DH5a. We designed a novel protocol to exam fluctuation of leu concentration in liquid LB medium. The protocol goes that:
Materials that should be prepared in advance
- 1 sulfosalicylic acid solution 20 g solid sulfosalicylic acid is dissolved in 100ml ddwater
- 2 ninhydrin solution 1 g ninhydrin is dissolved in 35ml ddwater
- 3 pH8.03 PBS
A solution 4.5350g KH2PO4 is dissolved in 500ml ddwater B solution 11.938g Na2HPO4 is dissolved in 500ml ddwater pH8.03 PBS = 10ml A solution +190ml B solution
procedure
- 1 1ml sterilized LB medium is inoculated with microbe in 1.5 ml EP tube and shaking in 37 ℃ overnight
- 2 10000g*1min for centrifugation and then pipette 800ul supernatant to another new 1.5 ml EP tube
- 3 pipette 300 ml LB to other two new 1.5 ml tubes and add 100ul sulfosalicylic acid solution
- 4 shaking violently for 1 min
- 5 20000g*10min for centrifugation and then pipette 300ul supernatant to a new 1.5 EP tube
- 6 add 1ml pH8.4 PBS buffer and mix them by reversing the tube several times
- 7 all 0.15ml ninhydrin solution and mix them by reversing the tube several times
- 8 incubate the tube in room temperature for 1hr
- 9 detect the optical density via spectrophotometer
full wavelength scanning
Before further test, we exam two kinds of liquid LB medium that are with or without additional leucine implementation. The liquid medium was processed follow the protocol above and tested by spectrophotometer for full wavelength scanning from 300nm to 1000nm. Since liquid LB medium teems with amino acid, short peptide and other nutrition, we can see a peak in B (without leucine implementation) at 570nm as well.
optical density at 570nm
The optical density as 570nm of liquid LB implemented with leucine. Nctr is a negative control set that do not contain microbe. 2101 is a negative control as well but inoculates with an E.coli named 2101 that harbors chloromycetin resistance but only expresses a short non-coding RNA. LivJKHMGF in the figure indicates medium after shaking overnight with E.coli DH5a contain corresponding overexpress vectors. The data of BL21 is not shown here since the data of different sets only owns tiny discrepency.
We try to explain these results, but we are not sure the following explanation reflects reality to what extent. To ascertain this question, there need more sophisticated investigation.
As we have mentioned before, liv JKHMGF are not resides on genome of Bl21. In addition, the leucine ABC transporter will not work if there is not other component expressed in cell. Thus, it can be understood that there are tiny discrepency among different sets of experiments in BL21.
For DH5a, Albeit the DH5a owns livJKHMGF in genome, the expressions of them are repressed by a regulator, Lrp (leucine-responsive regulator protein). According to the result of BLAST, Lrp not only resides on genome of DH5a but that of BL21. Lrp is a dual transcriptional regulator for at least 10% of the genes in Escherichia coli. These genes are involved in amino acid biosynthesis and catabolism, nutrient transport, pili synthesis, and other cellular functions. It is believed that Lrp senses the presence of rich nutrition based on the concentration of leucine and positively regulates genes that function during famine and negatively regulates genes that function during a feast.
Lrp-leucine DNA bingding transcriptional dual regulator inhibits livJ transcription initiation, whereas L-leucine binds to Lrp transcriptional dual regulator to block inhibition of livKHGMF.
As shows in figure 8, livJK seem do not increase uptake of leucine compared with 2101, livHM inhibit uptake of leucine compared with 2101 and livGF promote uptake of leucine dramatically, especially for livF, compared with 2101. We can brace these six genes into 3 group, livJK, livHM and livGF, according to the result of spectrophotometer. Then we attempt to ascertain the inner relationship that brace these genes into the 3 group.
explanation of optical density data
effect of leucine in coding region
Firstly, we list leucine number and percentage in these genes, and we considered that if there are more leucine in the protein, it will uptake more leucine from medium. But we did not find any relevance between these data and results in figure 8.
Then we scrutinize the construction of the leucine ABC transporter. Although there is a little information for the transporter, we find that the hydrophilicity plots can brace the six genes into three categories.
hydrophilicity plot of livJKHMGF
As figure 11,12,13 shows, livJK owns a hydrophobic N terminal. It is the signal peptide that guides livJ and livK to be transported through inner membrane and to be trapped in periplasmic space. livHM should be transmembrane protein since they have very hydrophobic peptide span. LivGF do not contain obvious hydrophobic peptide but contain Walker A and Walker B motifs involved in the binding and hydrolysis of ATP (according to results of BLAST). Thus we can imagine the structure of leucine ABC transporter which is shown in figure 14.
explanation for figure 8
Please note that all of those explanation of leucine uptake derived from conjecture. To ascertain the puzzle results, further and sophisticated experiment should be carried out.
Overexpress livJK in DH5a will not increase uptake of leucine in high leucine concentration medium (around 1ug/ml). Overexpress livMH in DH5a inhibits leucine uptake of leucine in high leucine concentration medium (around 1ug/ml). Overexpress livFG in DH5a promotes leucine uptake of leucine in high leucine concentration medium (around 1ug/ml). It may be explained that livFG works like a pump to hydrolysis ATP and obtain energy to transport leucine from periplasmic space into cytosol. More livFG means more energy is used to pump leucine into cytosol. Overexpress livHM may result in more transmembrane pathway which may be a handicap to retain leucine in cell cytosol without futher energy to determine direction of transportation, that is, there will be some leak of leucine when overexpress livHM. Overexpress livJK can not increase uptake of leucine may be explained that livJ and livK are saturated for transporter livHMGF, that is, more livJK to pass leucine to transporter livHMGF than the transporter needs. In addition, overexpressed livJK do not have enough time to be transported into periplasmic space and congregated together in cell and livJK do not have enough enzyme to cleave N-terminal of livJK, which may be another explanation for performance of overexpressed livJK. The two explanation for livJK are not mutual excluded.
Again,please note that all of those explanation of leucine uptake derived from conjecture. To ascertain the puzzle results, further and sophisticated experiment should be carried out.